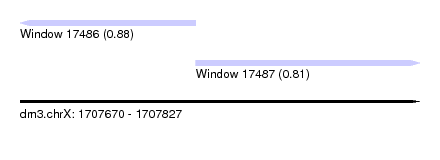
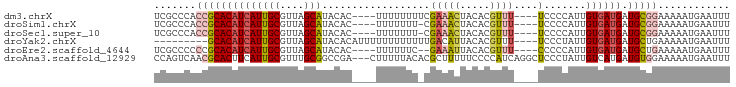
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,707,670 – 1,707,827 |
| Length | 157 |
| Max. P | 0.878509 |

| Location | 1,707,670 – 1,707,739 |
|---|---|
| Length | 69 |
| Sequences | 4 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 86.90 |
| Shannon entropy | 0.20552 |
| G+C content | 0.61617 |
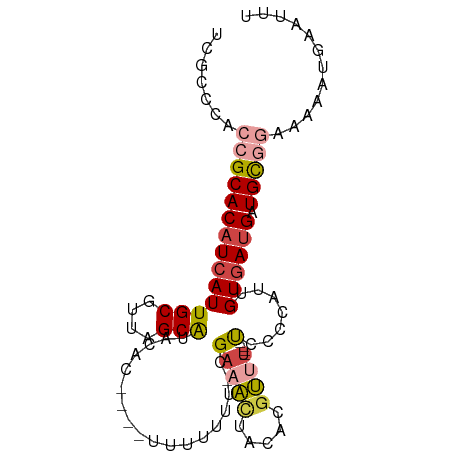
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -19.45 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
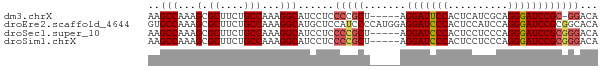
>dm3.chrX 1707670 69 - 22422827 AAGCCAAAGCGCUUCUGCCAAAGGCAUCCUCCCCGCU-----AGGAUUCCACUCAUCGCAGGGAUCCGC-GGACA ..(((...(.((....)))...))).......((((.-----.(((((((..........)))))))))-))... ( -21.50, z-score = -1.04, R) >droEre2.scaffold_4644 1688492 75 - 2521924 GUGCCAAAGCGCUUCUGCCAAAGGCAUGCUCCAUCCCCAUGGAGGAUCCCACUCCAUCCAGGGAUCCGCGGCACA (((((..........((((...)))).(((((((....)))))(((((((..........)))))))))))))). ( -31.90, z-score = -3.28, R) >droSec1.super_10 1502484 70 - 3036183 AAGCCAAAGCGCUUCUGCCAAAGGCAUCCUCCCCGCU-----AGGAUCCCACUCCUCCCAGGGAUCCGCGGGACA ..(((...(.((....)))...)))......(((((.-----.(((((((..........))))))))))))... ( -27.70, z-score = -2.69, R) >droSim1.chrX 1200849 70 - 17042790 AAGCCAAAGCGCUUCUGCCAAAGGCAUCCUCCCCGCU-----AGGAUCCCACUCCUCCCAGGGAUCCGCGGGACA ..(((...(.((....)))...)))......(((((.-----.(((((((..........))))))))))))... ( -27.70, z-score = -2.69, R) >consensus AAGCCAAAGCGCUUCUGCCAAAGGCAUCCUCCCCGCU_____AGGAUCCCACUCCUCCCAGGGAUCCGCGGGACA ..(((...(.((....)))...)))......(((((.......(((((((..........))))))))))))... (-19.45 = -20.13 + 0.69)
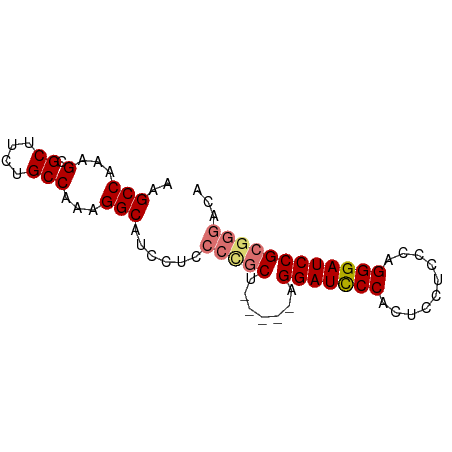
| Location | 1,707,739 – 1,707,827 |
|---|---|
| Length | 88 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Shannon entropy | 0.36124 |
| G+C content | 0.40336 |
| Mean single sequence MFE | -19.06 |
| Consensus MFE | -9.64 |
| Energy contribution | -10.58 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1707739 88 + 22422827 UCGCCCACCGCACAUCAUUGCGUUAGCAUACAC----UUUUUUUUCGAAACUACACGUUU----UCCCCAUUGUGAUGAUGCGGAAAAAUGAAUUU .......((((((((((((((....))).....----.........(((((.....))))----).......)))))).)))))............ ( -18.90, z-score = -2.22, R) >droSim1.chrX 1200919 87 + 17042790 UCGCCCACCGCACAUCAUUGCGUUAGCAUACAC----UUUUUUU-CGAAACUACACGUUU----UCCCCAUUGUGAUGAUGCGGAAAAAUGAAUUU .......((((((((((((((....))).....----.......-.(((((.....))))----).......)))))).)))))............ ( -18.90, z-score = -2.15, R) >droSec1.super_10 1502554 87 + 3036183 UCGCCCACCGCACAUCAUUGCGUUAGCAUACAC----UUUUUUU-CGAAACUACACGUUU----UCCCCAUUGUGAUGAUGCGGAAAAAUGAAUUU .......((((((((((((((....))).....----.......-.(((((.....))))----).......)))))).)))))............ ( -18.90, z-score = -2.15, R) >droYak2.chrX 1553290 83 + 21770863 ---------GCACAUCAUUGCGUUAGCAUACACAUUUUUUUUUUUUGACAUUACACGUUU----UCCCUAUUGUGAUGAUGCUGAAAAAUGAAUUU ---------.....(((((...(((((((...((...........)).(((((((.....----.......))))))))))))))..))))).... ( -15.70, z-score = -1.99, R) >droEre2.scaffold_4644 1688567 86 + 2521924 UCGCCCCCCGCACAUCAUUGCGUUAGCAUACAC----UUUUUUC--GAAAUUACACGUUU----CCCCCAUUGUGAUGAUGCUGAAAAAUGAAUUU ..((.....))...(((((...(((((((.(((----.......--(((((.....))))----).......)))...)))))))..))))).... ( -17.24, z-score = -2.73, R) >droAna3.scaffold_12929 1725897 93 - 3277472 CCAGUCAACGCACUUCAUUGCGUUUGCGGCCGA---CUUUUUACACGCUUUUUCCCCAUCAGGCUCCCUAUUGUCAUGAUGUGGAAAAAUGAAUUU ((.((.((((((......)))))).))))....---.........((.(((((((.((((((((........))).))))).)))))))))..... ( -24.70, z-score = -2.48, R) >consensus UCGCCCACCGCACAUCAUUGCGUUAGCAUACAC____UUUUUUU_CGAAACUACACGUUU____UCCCCAUUGUGAUGAUGCGGAAAAAUGAAUUU .......((((((((((((((....)))...................((((.....))))............)))))).)))))............ ( -9.64 = -10.58 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:30 2011