
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,680,602 – 1,680,693 |
| Length | 91 |
| Max. P | 0.836209 |

| Location | 1,680,602 – 1,680,693 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Shannon entropy | 0.41906 |
| G+C content | 0.33989 |
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.836209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
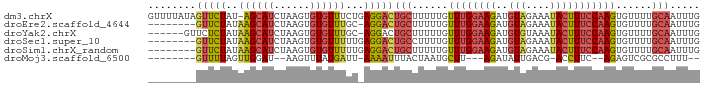
>dm3.chrX 1680602 91 - 22422827 GUUUUAUAGUUCUAU-AGCAUCUAAGUGUGUUUCUGAGGACUGCUUUUUGUUUGGAAGAUGUAGAAAUACUUUCCAAGUGUUUUGCAAUUUG .......((((((..-(((((......)))))....))))))((....(..(((((((..(((....))))))))))..)....))...... ( -19.80, z-score = -1.41, R) >droEre2.scaffold_4644 1658886 83 - 2521924 --------GUUCUAUAAGCAUCUAAGUGUGUUUGC-AGGACUGCUUUUUGUUUGGAAGAUGUAGAAAUACUUUCCAAGUGUUUUGCAAUUUG --------(((((.(((((((......))))))).-)))))(((....(..(((((((..(((....))))))))))..)....)))..... ( -22.10, z-score = -2.42, R) >droYak2.chrX 1525777 85 - 21770863 ------GUUCUCUAUAAGCAUCUAAGUGUGUUUGC-AGGACUGCUUUUUGUUUGGAAGAUGUGUAAAUACUUUCCAAGUGUUUUGCAAUUUG ------(((((...(((((((......))))))).-)))))(((....(..(((((((.(((....))).)))))))..)....)))..... ( -18.50, z-score = -1.14, R) >droSec1.super_10 1475469 84 - 3036183 --------GUUCUAUAAGCAUCUAAGUGUGUUUUUGAGGACUGCCUUUUGUUUGGAAGAUGUAGAAAUACUUUCCAAGUGUUUUGCAAUUUG --------(((((..((((((......))))))...)))))(((....(..(((((((..(((....))))))))))..)....)))..... ( -20.20, z-score = -2.11, R) >droSim1.chrX_random 1082388 84 - 5698898 --------GUUCUAUAAGCAUCUAAGUGUGUUUUUGAGGACUGCUUUUUGUUUGGAAGAUGUAGAAAUACUUUCCAAGUGUUUUGCAAUUUG --------(((((..((((((......))))))...)))))(((....(..(((((((..(((....))))))))))..)....)))..... ( -20.90, z-score = -2.36, R) >droMoj3.scaffold_6500 30845568 73 + 32352404 --------GUUUUAGUUGGAU--AAGUUUAUGAUU-AAAAUUUACUAAUGCUU---AGAUAUUGACG-ACCUUC--AGAGUCGCGCCUUU-- --------......(((((.(--((((((......-.))))))))))))((..---.(((.((((..-....))--)).)))..))....-- ( -9.90, z-score = 0.24, R) >consensus ________GUUCUAUAAGCAUCUAAGUGUGUUUUU_AGGACUGCUUUUUGUUUGGAAGAUGUAGAAAUACUUUCCAAGUGUUUUGCAAUUUG ........(((((..((((((......))))))...)))))(((......((((((((..(((....)))))))))))......)))..... (-12.00 = -12.80 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:21 2011