
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,672,738 – 1,672,828 |
| Length | 90 |
| Max. P | 0.991501 |

| Location | 1,672,738 – 1,672,828 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Shannon entropy | 0.31394 |
| G+C content | 0.51237 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.30 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1672738 90 + 22422827 CCAUCCCCAAGCACUAGGAUGUGCCCAAUGUGCCAUGCGGGCAUAUCCCAGAUUGCUAGAUCAAAUGCGAAUUA-GUACACUGAUUCCCAA- .........((((((.((((((((((.(((...)))..)))))))))).))..))))........((.((((((-(....))))))).)).- ( -30.50, z-score = -3.81, R) >droYak2.chrX 1518075 82 + 21770863 CCCUCUCGUAGCACUAGGAUGUGCCCAUUGGGCC----GGGCACAUUCCA--UUGCUAGUUCGAAUGCGAAUGAAGUCCACUGAUCCA---- ........(((((...((((((((((........----))))))))))..--.)))))(((((....)))))................---- ( -26.20, z-score = -1.36, R) >droEre2.scaffold_4644 1649523 86 + 2521924 CCCUCUCCUAGCACUAGGAUAUGCCCAUUGGGAC----GGGCAUGUCCCA--UUGCUAGAUCAAAUGCGAAAAGUGUCCACUGAUCCCCAAA .......((((((...((((((((((........----))))))))))..--.)))))).........((..(((....)))..))...... ( -28.60, z-score = -2.86, R) >consensus CCCUCUCCUAGCACUAGGAUGUGCCCAUUGGGCC____GGGCAUAUCCCA__UUGCUAGAUCAAAUGCGAAUAA_GUCCACUGAUCCCCAA_ .......((((((...((((((((((............)))))))))).....))))))................................. (-21.52 = -21.30 + -0.22)

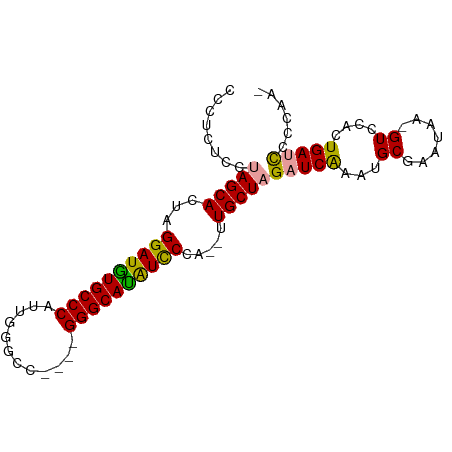
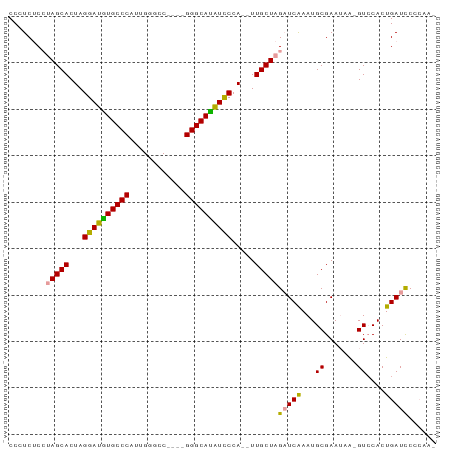
| Location | 1,672,738 – 1,672,828 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Shannon entropy | 0.31394 |
| G+C content | 0.51237 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -23.32 |
| Energy contribution | -25.43 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1672738 90 - 22422827 -UUGGGAAUCAGUGUAC-UAAUUCGCAUUUGAUCUAGCAAUCUGGGAUAUGCCCGCAUGGCACAUUGGGCACAUCCUAGUGCUUGGGGAUGG -.((.((((.((....)-).)))).))....((((((((..(((((((.((((((.(((...))))))))).)))))))))))...)))).. ( -30.30, z-score = -1.61, R) >droYak2.chrX 1518075 82 - 21770863 ----UGGAUCAGUGGACUUCAUUCGCAUUCGAACUAGCAA--UGGAAUGUGCCC----GGCCCAAUGGGCACAUCCUAGUGCUACGAGAGGG ----.......(((((.....))))).((((...(((((.--.(((.(((((((----(......)))))))))))...))))))))).... ( -29.40, z-score = -1.96, R) >droEre2.scaffold_4644 1649523 86 - 2521924 UUUGGGGAUCAGUGGACACUUUUCGCAUUUGAUCUAGCAA--UGGGACAUGCCC----GUCCCAAUGGGCAUAUCCUAGUGCUAGGAGAGGG ....(.((..(((....)))..)).)......(((((((.--(((((.((((((----((....)))))))).))))).)))))))...... ( -33.10, z-score = -2.59, R) >consensus _UUGGGGAUCAGUGGAC_UAAUUCGCAUUUGAUCUAGCAA__UGGGAUAUGCCC____GGCCCAAUGGGCACAUCCUAGUGCUAGGAGAGGG ........((.(((((.....)))))......(((((((...((((((((((((............)))))))))))).))))))).))... (-23.32 = -25.43 + 2.11)

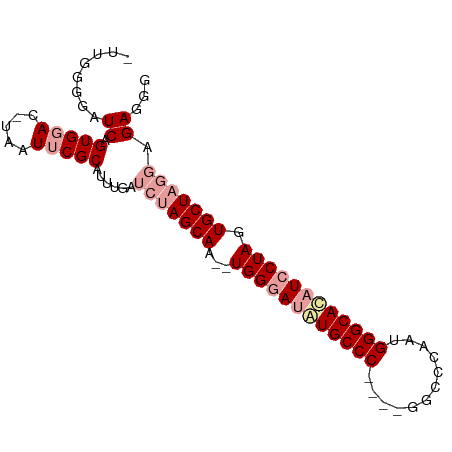
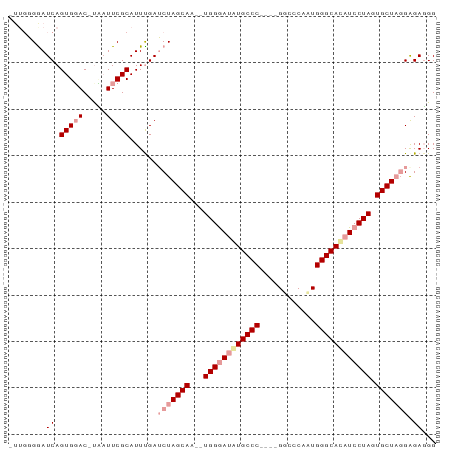
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:19 2011