| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,667,646 – 1,667,757 |
| Length | 111 |
| Max. P | 0.999524 |

| Location | 1,667,646 – 1,667,757 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 58.61 |
| Shannon entropy | 0.81992 |
| G+C content | 0.35331 |
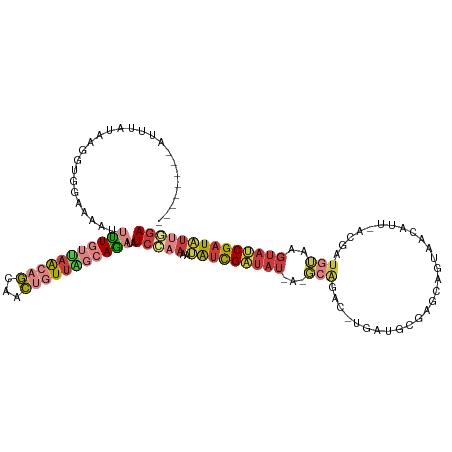
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -10.85 |
| Energy contribution | -12.52 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
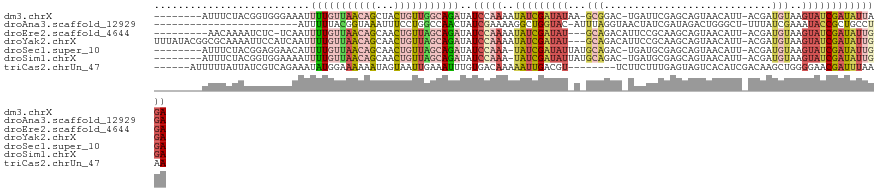
>dm3.chrX 1667646 111 + 22422827 --------AUUUCUACGGUGGGAAAUUUUGUUAACAGCUACUGUUGGCAGAUAUCCAAAAUAUCGAUAUAA-GCGGAC-UGAUUCGAGCAGUAACAUU-ACGAUGUAAGUAUCGAUAUUAGA --------............(((...(((((((((((...)))))))))))..)))..(((((((((((..-....((-((.......)))).((((.-...))))..)))))))))))... ( -29.00, z-score = -2.47, R) >droAna3.scaffold_12929 1673720 96 - 3277472 ------------------------AUUUUUACGGUAAAUUUCCUGGCCAACUAUCGAAAAGGCUGGUAC-AUUUAGGUAACUAUCGAUAGACUGGGCU-UUUAUCGAAAUACCGCUGCCUGA ------------------------........((((......(..(((............)))..)...-.....((((....((((((((.......-))))))))..))))..))))... ( -20.80, z-score = -0.41, R) >droEre2.scaffold_4644 1644664 108 + 2521924 ---------AACAAAAUCUC-UCAAUUUUGUUAACAGCAACUGUUAGCAGAUAUCCAAAAUAUCGAUAU---GCAGACAUUCCGCAAGCAGUAACAUU-ACGAUGUAAGUAUCGAUAUUGGA ---------...........-.....(((((((((((...)))))))))))..((((..((((((((((---....(((((.(....)..(((....)-)))))))..)))))))))))))) ( -28.80, z-score = -4.30, R) >droYak2.chrX 1512840 118 + 21770863 UUUAUACGGCGCAAAAUUCCAUCAAUUUUGUUAACAGCAACUGUUAGCAGAUAUCCAAAAUAUCGAUAU---GCAGACAUUCCGCAAGCAGUAACAUU-ACGAUGUAAGUAUCGAUAUUGGA .................(((......(((((((((((...))))))))))).......(((((((((((---....(((((.(....)..(((....)-)))))))..)))))))))))))) ( -28.90, z-score = -3.13, R) >droSec1.super_10 1461948 111 + 3036183 --------AUUUCUACGGAGGAACAUUUUGUUAACAGCAACUGUUAGCAGAUAUCCAAA-UAUCGAUAUUAUGCAGAC-UGAUGCGAGCAGUAACAUU-ACGAUGUAAGUAUCGAUAUUGGA --------.((((....)))).....(((((((((((...)))))))))))..(((((.-((((((((((.((((...-(((((..........))))-)...))))))))))))))))))) ( -33.40, z-score = -4.28, R) >droSim1.chrX 1176673 111 + 17042790 --------AUUUCUACGGUGGAAAAUUUUGUUAACAGCAACUGUUAGCAGAUAUCCAAA-UAUCGAUAUUAUGCAGAC-UGAUGCGAGCAGUAACAUU-ACGAUGUAAGUAUCGAUAUUGGA --------.(((((.....)))))..(((((((((((...)))))))))))..(((((.-((((((((((.((((...-(((((..........))))-)...))))))))))))))))))) ( -32.40, z-score = -3.66, R) >triCas2.chrUn_47 33034 108 - 241962 ------AUUUUUAUUAUCGUCAGAAAUAUGGAAAAAAUAGUAAUUGAAAUUUGUGACAAAAAUUGACGU--------UCUUCUUUGAGUAGUCACAUCGACAAGCUGGGGAACGAUUUAAAA ------............((((.((((.(.((...........)).).)))).)))).........(((--------(((((...(..(.(((.....))))..).))))))))........ ( -17.30, z-score = 0.24, R) >consensus ________AUUUAUAAGGUGGAAAAUUUUGUUAACAGCAACUGUUAGCAGAUAUCCAAAAUAUCGAUAU_A_GCAGAC_UGAUGCGAGCAGUAACAUU_ACGAUGUAAGUAUCGAUAUUGGA ..........................(((((((((((...)))))))))))..(((((..(((((((((...(((............................)))..)))))))))))))) (-10.85 = -12.52 + 1.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:18 2011