
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,658,580 – 1,658,682 |
| Length | 102 |
| Max. P | 0.756689 |

| Location | 1,658,580 – 1,658,682 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.69 |
| Shannon entropy | 0.28077 |
| G+C content | 0.39606 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.37 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
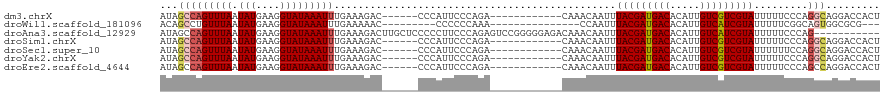
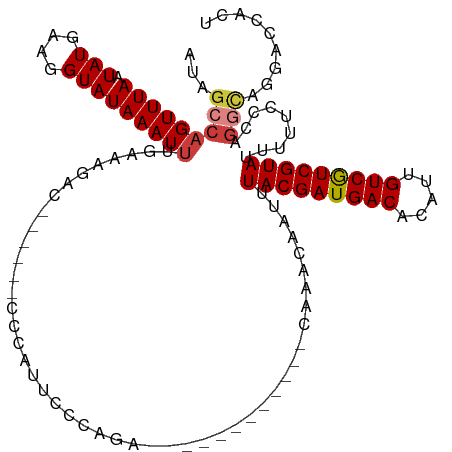
>dm3.chrX 1658580 102 + 22422827 AUAGCCAGUUUAAUAUGAAGGUAUAAAUUUGAAAGAC------CCCAUUCCCAGA------------CAAACAAUUUACGAUGACACAUUGUCGUCGUAUUUUUCCCAGGCAGGACCACU ...(((.((((...(((..(((.............))------).)))....)))------------)........(((((((((.....))))))))).........)))......... ( -20.92, z-score = -2.09, R) >droWil1.scaffold_181096 4594731 93 + 12416693 ACAGCCUGUUUAAUAUGAAGGUAUAAAUUUGAAAAAC---------CCCCCCAAA---------------CCAAUUUACGAUGACACAUUGUCAUCGUAUUUUUCGGCAGUGGCGCG--- ...((((...........))))...............---------....(((..---------------((((..(((((((((.....)))))))))..))..))...)))....--- ( -20.00, z-score = -1.46, R) >droAna3.scaffold_12929 1535490 109 + 3277472 AUAGCCAGUUUAAUAUGAAGGUAUAAAUUUGAAAGACUUGCUCCCCCUUCCCAGAGUCCGGGGGAGACAAACAAUUUACGAUGACACAUUGUCAUCGUAUUUUUCCCAG----------- ...(((.............)))........((((((.((((((((((............))))))).)))......(((((((((.....)))))))))))))))....----------- ( -31.62, z-score = -3.60, R) >droSim1.chrX 1167466 102 + 17042790 AUAGCCAGUUUAAUAUGAAGGUAUAAAUUUGAAAGAC------CCCAUUCCCAGA------------CAAACAAUUUACGAUGACACAUUGUCGUCGUAUUUUUCCCAGGCAGGACCACU ...(((.((((...(((..(((.............))------).)))....)))------------)........(((((((((.....))))))))).........)))......... ( -20.92, z-score = -2.09, R) >droSec1.super_10 1453250 102 + 3036183 AUAGCCAGUUUAAUAUGAAGGUAUAAAUUUGAAAGAC------CCCAUUCCCAGA------------CAAACAAUUUACGAUGACACAUUGUCGUCGUAUUUUUUCCAGGCAGGACCACU ...(((.((((...(((..(((.............))------).)))....)))------------)........(((((((((.....))))))))).........)))......... ( -20.92, z-score = -2.09, R) >droYak2.chrX 1503525 102 + 21770863 AUAGCCAGUUUAAUAUGAAGGUAUAAAUUUGAAAGAC------CCCAUUCCCAGA------------CAAACAAUUUACGAUGACACAUUGUCGUCGUAUUUUUCCCAGGCAGGACCACU ...(((.((((...(((..(((.............))------).)))....)))------------)........(((((((((.....))))))))).........)))......... ( -20.92, z-score = -2.09, R) >droEre2.scaffold_4644 1634806 102 + 2521924 AUAGCCAGUUUAAUAUGAAGGUAUAAAUUUGAAAGAC------CCCAUUCCCAGA------------CAAACAAUUUACGAUGACACAUUGUCGUCGUAUUUUUCCCAGCCAGGACCACU ....((.((((...(((..(((.............))------).)))....)))------------)........(((((((((.....))))))))).............))...... ( -16.92, z-score = -1.36, R) >consensus AUAGCCAGUUUAAUAUGAAGGUAUAAAUUUGAAAGAC______CCCAUUCCCAGA____________CAAACAAUUUACGAUGACACAUUGUCGUCGUAUUUUUCCCAGGCAGGACCACU ...(((((((((.(((....)))))))))...............................................(((((((((.....))))))))).........)))......... (-13.96 = -14.37 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:16 2011