| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,655,833 – 1,655,925 |
| Length | 92 |
| Max. P | 0.734892 |

| Location | 1,655,833 – 1,655,925 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 67.01 |
| Shannon entropy | 0.59078 |
| G+C content | 0.47017 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -11.04 |
| Energy contribution | -9.89 |
| Covariance contribution | -1.15 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
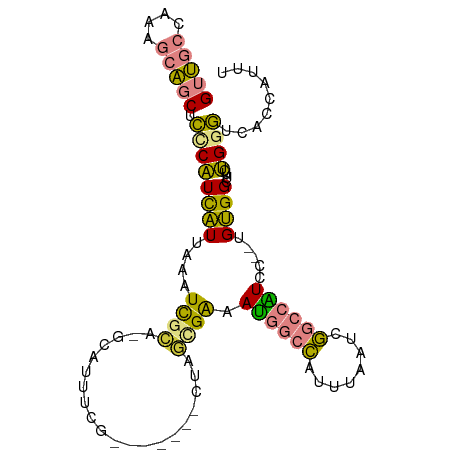
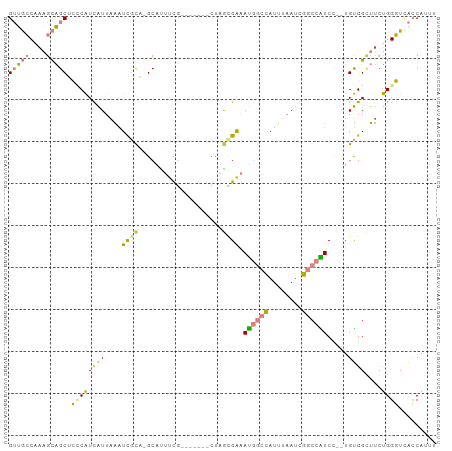
>dm3.chrX 1655833 92 + 22422827 GUUGCCAAAGCAGCACCCAUCAUUAAAUCGCU-GCAUUUCG-------CUUGCGAAAUGGCCAUUUAAUCGGCCAUCC--UGUGGCUUCUGGGUCACCAUUU (((((....)))))(((((..(((((((.((.-.(((((((-------....))))))))).))))))).((((((..--.))))))..)))))........ ( -30.30, z-score = -2.64, R) >droSim1.chrX 1164818 92 + 17042790 GUUGCCAAAGCAGCUCCCAUCAUUAAAUCGCA-GCACUUCG-------CUAGCGAAAUGGCCAUUUAAUCGGCCAUCC--UGUGGCUUCUGGGUCACCAUUU (((((....))))).............(((((-((.....)-------)).)))).((((((........))))))..--.((((((....))))))..... ( -28.90, z-score = -2.54, R) >droSec1.super_10 1450689 92 + 3036183 GUUGCCAAAGCAGCUCCCAUCAUUAAAUCGCA-GCACUUCG-------CUAGCGAAAUGGCCAUUUAAUCGGCCAUCC--UGUGGCUUCUGGGUCACCAUUU (((((....))))).............(((((-((.....)-------)).)))).((((((........))))))..--.((((((....))))))..... ( -28.90, z-score = -2.54, R) >droYak2.chrX 1499165 98 + 21770863 GUUGCCAAAGCAGCUCACAUCAUUAAAUCGCA-GCAUUUCGUAGCAGUCAAAUGGAAUGGCCAUUUAAUCGGCCAUCC--UGUGGCUUCUGGGUCACCAUU- (((((....)))))...............((.-((.....)).))(((((...(((.(((((........))))))))--..)))))..(((....)))..- ( -28.20, z-score = -1.90, R) >droEre2.scaffold_4644 1632555 92 + 2521924 GUUGCCAAAGCAGCUCACAUCAUUAAAUCGCA-GCACUGCG-------CCAGUGAAAUGGCCAUUUAAUCGGCCGUCC--UGUGGGUUCUGGGUCACCAAUA ((.(((..((.(((((((((((((....((((-....))))-------..))))).((((((........))))))..--)))))))))).))).))..... ( -29.50, z-score = -1.65, R) >dp4.chrXL_group1a 4832709 101 - 9151740 GAGUACAAUGUUGCAAUCGUUGUUAGGUUUUUUGCAUAUUUUCAGCG-AUUACGACACAAAAAUUGAAUCUCCAGUCGAAUGCGGUCCUCGGUGUGGCAUUG .....((((((..(((((((((....((.....)).......)))))-))).((((..................))))...............)..)))))) ( -21.07, z-score = 0.48, R) >droPer1.super_11 1949331 101 - 2846995 GAGUACAAUGUUGCAAUCGUUGUUAGGUUUUUUGCAUAUUUUCAGCG-AUUACGACACAAAAAUUGAAUCUCCAGUCGAAUGCGGUCUUCGGUGUGGCAUUG .....((((((..(((((((((....((.....)).......)))))-)))...........((((((..((((......)).))..)))))))..)))))) ( -21.80, z-score = 0.16, R) >consensus GUUGCCAAAGCAGCUCCCAUCAUUAAAUCGCA_GCAUUUCG_______CUAGCGAAAUGGCCAUUUAAUCGGCCAUCC__UGUGGCUUCUGGGUCACCAUUU (((((....))))).((((((((....((((....................)))).((((((........)))))).....))))....))))......... (-11.04 = -9.89 + -1.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:15 2011