| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,655,176 – 1,655,272 |
| Length | 96 |
| Max. P | 0.549586 |

| Location | 1,655,176 – 1,655,272 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.67009 |
| G+C content | 0.41176 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -7.14 |
| Energy contribution | -6.83 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1655176 96 - 22422827 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUCC--------GCUCCCAGAGUGGAGGUAUUGUGCCAUUUUUGGCGCUACU-CCUGCCACGUU--------- .....((((..(((..((((((.....))))))......((((((--------((((...))))))))))...((((((....))))))...)-)).))))....--------- ( -36.10, z-score = -4.19, R) >droSim1.chrX 1164162 96 - 17042790 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUCC--------GCUCCCAGAGUGGAGGUAUUGUGCCAUUUUUGGCGCUGCU-CCUGCCACGUU--------- .....((((..(((..((((((.....))))))......((((((--------((((...))))))))))...((((((....))))))...)-)).))))....--------- ( -36.10, z-score = -3.82, R) >droSec1.super_10 1450033 96 - 3036183 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUCC--------GCUCCCAGAGUGGAGGUAUUGUGCCAUUUUUGGCGCUGCU-CCUGCCACGUU--------- .....((((..(((..((((((.....))))))......((((((--------((((...))))))))))...((((((....))))))...)-)).))))....--------- ( -36.10, z-score = -3.82, R) >droYak2.chrX 1498490 100 - 21770863 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCCCCAAU----GGAGGUGGAGGUGGAGGUAUUGUGCCAUUUUUGGCGCUGCU-CCUGCCACGUC--------- ......(((((....)))))...............(((((((((....----)).)))))))(((((((....((((((....))))))....-))).))))...--------- ( -34.90, z-score = -2.08, R) >droEre2.scaffold_4644 1631932 94 - 2521924 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUAC----------GCUCGCAGUGGAGGCAUUGUCCCAUUUUUGGCGCUGCU-CCCGCCACGUU--------- ..((((((..((((((((((((.....)))))).)))))((((.(----------((.....))).))))...)..)))))).(((((.....-..)))))....--------- ( -28.20, z-score = -1.15, R) >droAna3.scaffold_12929 1531313 104 - 3277472 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUUCUCUCUGUCUGCCCCUCUUUGGAAAUAUUGUGCCAUUUUUGCCUUCGGU-CCUUUCCCGUU--------- ..(((((((.((((((((((((.....)))))).)))))).................((.....)).........))))))).......(((.-......)))..--------- ( -15.70, z-score = 0.51, R) >dp4.chrXL_group1a 4831904 88 + 9151740 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUCCUGCCU-----------CUGAAAAAUAUUGUGCCUUUU-----UUUGGU-CUUUUCACGUU--------- ((((((..((((((..((((((.....))))))......((.....)).)-----------)))))..)).))))(((....-----...)))-...........--------- ( -12.60, z-score = 0.47, R) >droPer1.super_11 1948526 88 + 2846995 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUCCUGCCU-----------CUGAAAAAUAUUGUGCCUUUU-----UUUGGU-CUUUUCACGUU--------- ((((((..((((((..((((((.....))))))......((.....)).)-----------)))))..)).))))(((....-----...)))-...........--------- ( -12.60, z-score = 0.47, R) >droWil1.scaffold_181096 4590787 100 - 12416693 ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUCU------------UGUCUGUGAAAAUAUUGUGCCAUUU--GCCAUUGGUACUUGGUGCUUUUCGCCCUUU ((((..(((..((((.((((((.....)))))).)))).)))..)------------)))..(((((((....(..(((..(--(((...))))..)))..))))))))..... ( -27.50, z-score = -3.11, R) >droVir3.scaffold_9476 44841 82 + 69990 ACAAAUGGUUUGGAAGAAUUGCUUUAAGUAAUUGUUUUUAUAUCU------------CUUCUAUGGACGUUUUGUGAUACUUUACCGUUGUUCU-------------------- ...((((((..((((.((((((.....)))))).)).(((((...------------(.((....)).)...)))))..))..)))))).....-------------------- ( -12.70, z-score = -0.49, R) >droMoj3.scaffold_6328 4358146 73 - 4453435 ACAAAUGGCGUGGAAGAAUUGCUUUAAGUAAUUGUUUUCCUAUCU------------CCUCUAUGAACAUUUUCUGGUUGUCCUU----------------------------- ((((..((.(.(((((((((((.....)))))..))))))...).------------)).(((.(((....))))))))))....----------------------------- ( -10.20, z-score = 0.18, R) >droGri2.scaffold_14853 757876 90 - 10151454 ACAAAUGGUUUGGAAGAAUUGCUUUAAGUAAUUGUUUUUCCAUCU------------CUUCUA-GAACA-UUUGUGAUAUUUUUUUCAGCCUGUCCCUGCUGUU---------- (((((((.(((((((((..((...(((....)))......))..)------------))))))-)).))-)))))...........((((........))))..---------- ( -20.30, z-score = -2.51, R) >consensus ACAAAUGGUUCGGAAGAAUUGCUUUAAGUAAUUGUUUUCGCCUCC____________CCUCUGUGGAGAUAUUGUGCCAUUUUUGGCGCUGCU_CCUGCCACGUU_________ ......((..((((((((((((.....)))))).))))))...))..................................................................... ( -7.14 = -6.83 + -0.30)

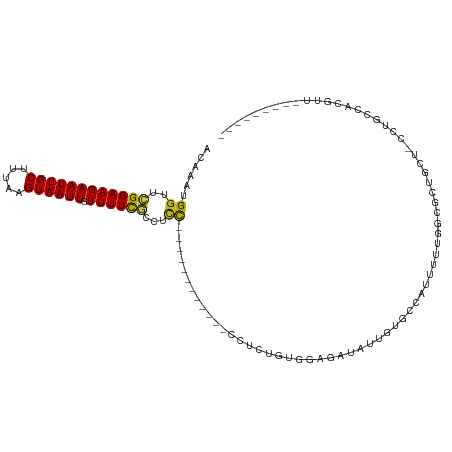

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:14 2011