
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,629,215 – 1,629,309 |
| Length | 94 |
| Max. P | 0.961615 |

| Location | 1,629,215 – 1,629,309 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.68 |
| Shannon entropy | 0.05861 |
| G+C content | 0.63176 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -29.72 |
| Energy contribution | -29.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
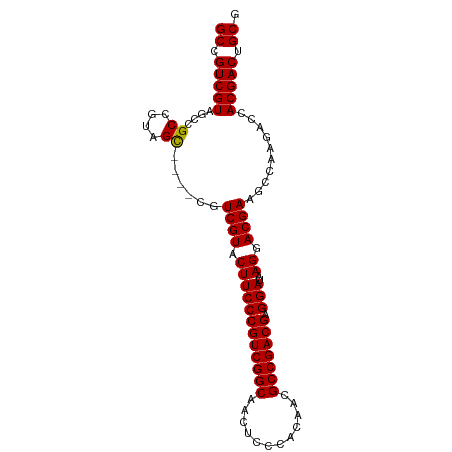
>dm3.chrX 1629215 94 + 22422827 GCCGUCGUAGCCGCCGUAGUAACUCAUCGUACUUCCCGUCGGCAACUCCCACAACGCCGACGAGGAUAAGGACGAAGCCAAGACCACGACUGCG ((.(((((.......((....))...((((.((((((((((((............))))))).)))..)).))))..........))))).)). ( -29.30, z-score = -1.81, R) >droSim1.chrU 3926138 90 - 15797150 GCCGUCGUAGCCGCCGUAGC----CGUCGUACUUCCCGUCGGCAACUCCCACAACGCCGACGAGGAUAAGGACGAAGCCAAGACCACGACUGCG ((.(((((....((....))----((((.(...((((((((((............))))))).)))..).))))...........))))).)). ( -32.20, z-score = -2.20, R) >droSec1.super_10 1433587 90 + 3036183 GCCGUCGUAGCCGCCGUAGC----CGUCGUACUUCCCGUCGGCAACUCCCACAACGCCGACGAGGAUAAGGACGAAGCCAAGACCACGACUGCG ((.(((((....((....))----((((.(...((((((((((............))))))).)))..).))))...........))))).)). ( -32.20, z-score = -2.20, R) >consensus GCCGUCGUAGCCGCCGUAGC____CGUCGUACUUCCCGUCGGCAACUCCCACAACGCCGACGAGGAUAAGGACGAAGCCAAGACCACGACUGCG ((.(((((....((....))......((((.((((((((((((............))))))).)))..)).))))..........))))).)). (-29.72 = -29.50 + -0.22)

| Location | 1,629,215 – 1,629,309 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.68 |
| Shannon entropy | 0.05861 |
| G+C content | 0.63176 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -39.83 |
| Energy contribution | -40.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
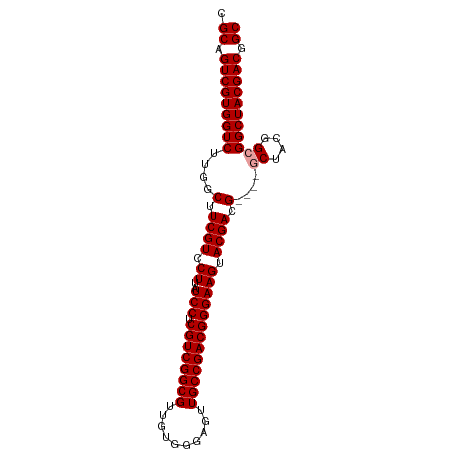
>dm3.chrX 1629215 94 - 22422827 CGCAGUCGUGGUCUUGGCUUCGUCCUUAUCCUCGUCGGCGUUGUGGGAGUUGCCGACGGGAAGUACGAUGAGUUACUACGGCGGCUACGACGGC .((.(((((((((.(((((((((.((..(((.((((((((..........))))))))))))).))))..)))))(....).))))))))).)) ( -40.10, z-score = -1.83, R) >droSim1.chrU 3926138 90 + 15797150 CGCAGUCGUGGUCUUGGCUUCGUCCUUAUCCUCGUCGGCGUUGUGGGAGUUGCCGACGGGAAGUACGACG----GCUACGGCGGCUACGACGGC .((.(((((((((.(((((((((.((..(((.((((((((..........))))))))))))).)))).)----))))....))))))))).)) ( -42.50, z-score = -2.21, R) >droSec1.super_10 1433587 90 - 3036183 CGCAGUCGUGGUCUUGGCUUCGUCCUUAUCCUCGUCGGCGUUGUGGGAGUUGCCGACGGGAAGUACGACG----GCUACGGCGGCUACGACGGC .((.(((((((((.(((((((((.((..(((.((((((((..........))))))))))))).)))).)----))))....))))))))).)) ( -42.50, z-score = -2.21, R) >consensus CGCAGUCGUGGUCUUGGCUUCGUCCUUAUCCUCGUCGGCGUUGUGGGAGUUGCCGACGGGAAGUACGACG____GCUACGGCGGCUACGACGGC .((.(((((((((......((((.((..(((.((((((((..........))))))))))))).))))......((....))))))))))).)) (-39.83 = -40.17 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:07 2011