
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,628,037 – 1,628,135 |
| Length | 98 |
| Max. P | 0.510658 |

| Location | 1,628,037 – 1,628,135 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.75 |
| Shannon entropy | 0.44914 |
| G+C content | 0.38932 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -10.05 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1628037 98 - 22422827 -----------CUUAAGUGCCAUGGUAG-GGGGGGUGUGGCAUUUC----UUUCA-CGUUGCAGUUAAUAGGAUUUGGCUGA---AGAGCGGAAUGAAAUUAAUUGUUUCAUUUGUUU -----------...((((((((((.(..-....).)))))))))).----.....-((((.(((((((......))))))).---..))))(((((((((.....))))))))).... ( -26.90, z-score = -2.24, R) >droYak2.chrX 1480061 94 - 21770863 ------------UUCAA----GUGCUGAGGGGGUGAAAGGCAUUUC----UUUCA-CGUUGCAGUUAAUAGGAUUUGGCUGG---AGAGUGGAAUGAAAUUAAUUGUUUCAUUUGUUU ------------.....----.....((((..(((.....)))..)----)))((-(.((.(((((((......))))))).---)).)))(((((((((.....))))))))).... ( -24.30, z-score = -2.02, R) >droEre2.scaffold_4644 1615085 93 - 2521924 ------------CUCAA----GUGGCGA-GGGGUGAAAGGCAUUUC----UUUCA-CGUUGCAGUUAAUAGGAUUUGGCUGG---AGAGUGGAACGAAAUUAAUUGUUUCAUUUGUUU ------------(((..----((((.((-(..(((.....)))..)----)))))-)....(((((((......))))))))---))((((((((((......))))))))))..... ( -27.10, z-score = -2.98, R) >droAna3.scaffold_12929 1513016 92 - 3277472 ------------CCCAA----GUGCCC--GUGGGAGAAAGCAUUUC----UUUUA-CGUUGCAGUUAAUAGGAUUUGGCUGG---AGAACGGAAUCAAAUUAAUUGUUUCAUUUGUUU ------------..(((----(((..(--(((((((((.....)))----)))))-))..(((((((((..(((((.(....---....).)))))..)))))))))..))))))... ( -24.60, z-score = -1.73, R) >dp4.chrXL_group1a 4809097 102 + 9151740 -----------CCCCAAAGGCAUGAAAAGGGGGAGAGAAACAUUUC----UUUAA-CGUUGCAGUUAAUAGGAUUUGGCUGAAACAGAGUGGAAUCAAAUUAAUUGUUUCAUUUGUUU -----------((((.............)))).((((((....)))----)))..-.(..(((((((((..(((((.(((.......))).)))))..)))))))))..)........ ( -25.52, z-score = -1.82, R) >droPer1.super_11 1925631 102 + 2846995 -----------CCCCAAAGGCAUGAAAAGGGGGAGAGAAACAUUUC----UUUAA-CGUUGCAGUUAAUAGGAUUUGGCUGAAACAGAGUGGAAUCAAAUUAAUUGUUUCAUUUGUUU -----------((((.............)))).((((((....)))----)))..-.(..(((((((((..(((((.(((.......))).)))))..)))))))))..)........ ( -25.52, z-score = -1.82, R) >droWil1.scaffold_181096 4564757 115 - 12416693 UGCGACAGAGAUGGAAACAUUGAGAAAGAGAAGGAGACCAGAUUCUCAGGUCUCACCGUUGCAGUUAAUAGGAUUUGACUGG---AGCCCGGAAUCAAAUUAAUUGUUUCAUUUGUUU ...((((((((((....))))..((((.((...((((((.(.....).)))))).(((((.(((((((......))))))).---)))..))...........)).)))).)))))). ( -32.30, z-score = -2.12, R) >consensus ____________CCCAA____GUGAAAA_GGGGGGAAAAGCAUUUC____UUUCA_CGUUGCAGUUAAUAGGAUUUGGCUGG___AGAGUGGAAUCAAAUUAAUUGUUUCAUUUGUUU .........................................................(..(((((((((...((((.(((.......))).))))...)))))))))..)........ (-10.05 = -10.60 + 0.55)

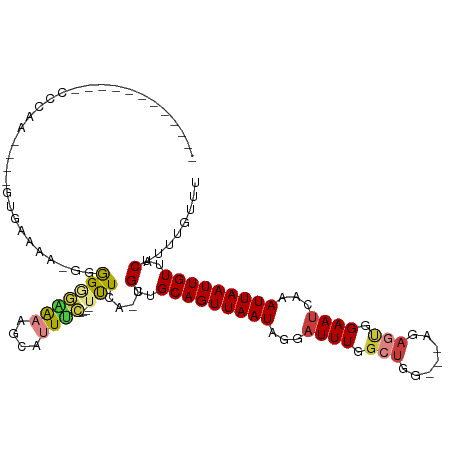
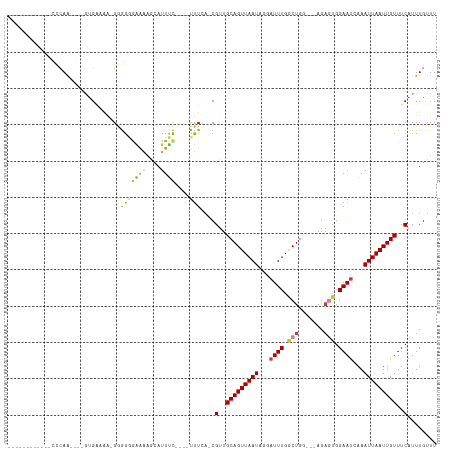
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:06 2011