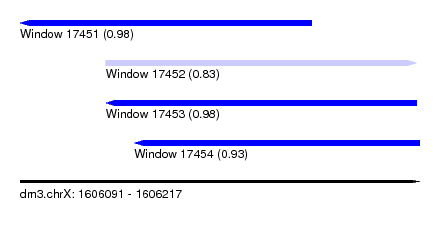
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,606,091 – 1,606,217 |
| Length | 126 |
| Max. P | 0.983851 |

| Location | 1,606,091 – 1,606,183 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.35 |
| Shannon entropy | 0.27478 |
| G+C content | 0.49436 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -22.45 |
| Energy contribution | -24.01 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
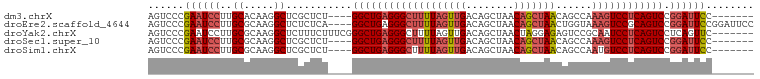
>dm3.chrX 1606091 92 - 22422827 GAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCCGG--------------AUUUUGGAUUCCGGAGUCUGCAGUCUGUUGCUUG--- (((((((((..((((..(((.....)))..)))).))))))))).....(((((((((--------------(........))))))))))((((....))))...--- ( -37.20, z-score = -3.81, R) >droEre2.scaffold_4644 1594109 109 - 2521924 GAGGGCUUUUAGUUGACAGCUAACAGCUAACUGGUAAAGUCCGCAGUCCGGAUUCCGGAUUCCGGAUUUUGCAUUUUGCAUUCCGGACUCUGCAGUCUGUUGCUUGAAU ...((((.(((((.....))))).)))).....((((((((((.(((((((...))))))).)))))))))).((..(((...((((((....)))))).)))..)).. ( -41.10, z-score = -3.57, R) >droYak2.chrX 1458757 95 - 21770863 GAGGGCUUUUAGUUGACAGCUAACUAGGAGAGUCCGCAAUCCUCAGUCCUCAGUUCGG--------------AAUUUGGAUUCCGGAGUCUGCAGUCUGUUGCUUGGAU ............(..(((((..(((.(.(((.((((.(((((....(((.......))--------------)....))))).)))).))).))))..)))).)..).. ( -29.50, z-score = -0.55, R) >droSec1.super_10 1410654 92 - 3036183 GAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCCGG--------------AUUUUGGAUUCCGGAGUCUGCAGUCUGUUGCUUG--- (((((((((..((((..(((.....)))..)))).))))))))).....(((((((((--------------(........))))))))))((((....))))...--- ( -37.20, z-score = -3.81, R) >droSim1.chrX 1137358 92 - 17042790 GAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAUGUCCUCAGUCCGGAUUCCGG--------------AUUUUGGAUUCCGGAGUCUGCAGUCUGUUGCUUG--- ...((((.(((((((........))))))).))))...((...(((.(((((((((((--------------(........)))))))))))..).)))..))...--- ( -34.50, z-score = -3.02, R) >consensus GAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCCGG______________AUUUUGGAUUCCGGAGUCUGCAGUCUGUUGCUUG___ (((((((((((((((........)))))))......))))))))..((((((.(((((.................))))).))))))....((((....))))...... (-22.45 = -24.01 + 1.56)

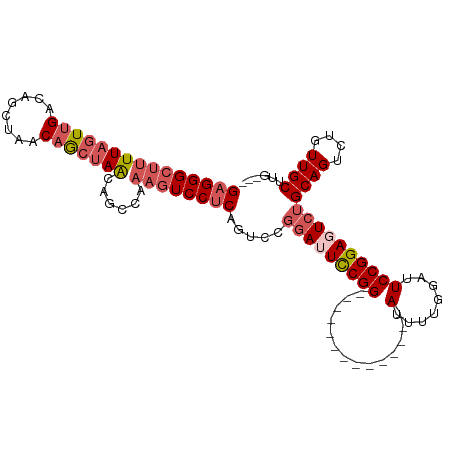
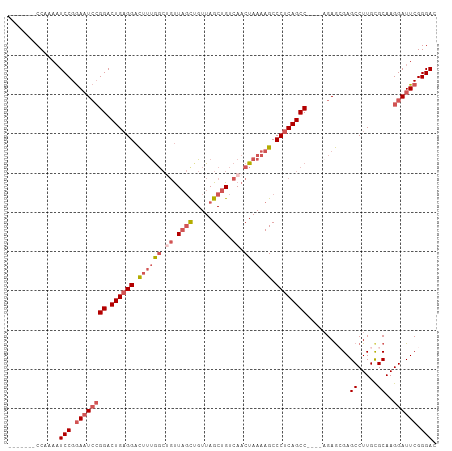
| Location | 1,606,118 – 1,606,216 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Shannon entropy | 0.25330 |
| G+C content | 0.53304 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -25.20 |
| Energy contribution | -27.52 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1606118 98 + 22422827 -------CCAAAAUCCGGAAUCCGGACUGAGGACUUUGGCUGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCC----AGAGCGAGCCUUGUGCAAGGAUUCGGGAC -------......(((.(((((((.((.((((.(((((((((...(((.((((.......)))).)))...)))))----))))....)))))).)..)))))).))). ( -39.00, z-score = -3.21, R) >droEre2.scaffold_4644 1594146 105 + 2521924 GCAAAAUCCGGAAUCCGGAAUCCGGACUGCGGACUUUACCAGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCC----UGAGAGAGCCUUGCGCAAGGAUUCGGGAC .............(((.((((((....(((((.((((..(((...((((...(((..........)))...)))))----))..))))))..)))...)))))).))). ( -31.00, z-score = -0.40, R) >droYak2.chrX 1458787 102 + 21770863 -------CCAAAUUCCGAACUGAGGACUGAGGAUUGCGGACUCUCCUAGUUAGCUGUCAACUAAAAGCCCUCAGCCCGAAAGAAAGAGCCUUGCGCAAGGAUUCGGGAC -------.....(((((((((..((.((((((...((((.....))(((((.......)))))...)))))))).))...))......((((....)))).))))))). ( -28.90, z-score = -0.64, R) >droSec1.super_10 1410681 98 + 3036183 -------CCAAAAUCCGGAAUCCGGACUGAGGACUUUGGCUGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCC----AGAGCGAGCCUUGCGCAAGGAUUCGGGAC -------......(((.((((((((.((((((.((((((.((.((((.....)))).)).)).)))).))))))))----...(((.......)))..)))))).))). ( -40.10, z-score = -3.33, R) >droSim1.chrX 1137385 98 + 17042790 -------CCAAAAUCCGGAAUCCGGACUGAGGACAUUGGCUGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCC----AGAGCGAGCCUUGCGCAAGGAUUCGGGAC -------......(((.((((((((.((((((...((((.((.((((.....)))).)).))))....))))))))----...(((.......)))..)))))).))). ( -37.90, z-score = -2.88, R) >consensus _______CCAAAAUCCGGAAUCCGGACUGAGGACUUUGGCUGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCC____AGAGCGAGCCUUGCGCAAGGAUUCGGGAC .............(((.((((((((.((((((.((((((.((.((((.....)))).)).)).)))).))))))))...........((.....))..)))))).))). (-25.20 = -27.52 + 2.32)

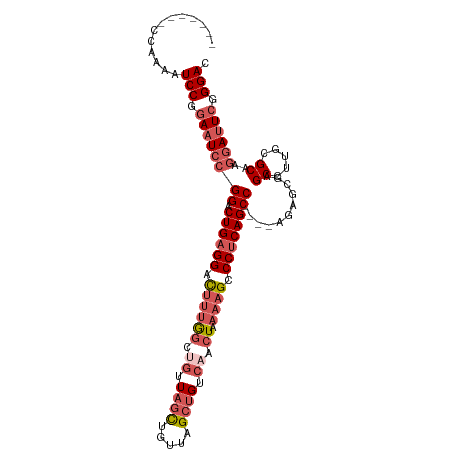
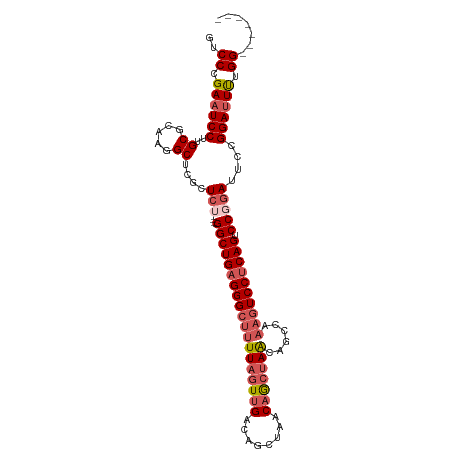
| Location | 1,606,118 – 1,606,216 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Shannon entropy | 0.25330 |
| G+C content | 0.53304 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -28.56 |
| Energy contribution | -30.28 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1606118 98 - 22422827 GUCCCGAAUCCUUGCACAAGGCUCGCUCU----GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCCGGAUUUUGG------- ((((.((((((..((.....)).......----(((((((((((((..((((..(((.....)))..)))).))))))))))))).)))))).)))).....------- ( -39.80, z-score = -3.76, R) >droEre2.scaffold_4644 1594146 105 - 2521924 GUCCCGAAUCCUUGCGCAAGGCUCUCUCA----GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACUGGUAAAGUCCGCAGUCCGGAUUCCGGAUUCCGGAUUUUGC .................(((((((((...----....)))))))))((((((........))))))....((((((((((.(((((((...))))))).)))))))))) ( -37.70, z-score = -1.82, R) >droYak2.chrX 1458787 102 - 21770863 GUCCCGAAUCCUUGCGCAAGGCUCUUUCUUUCGGGCUGAGGGCUUUUAGUUGACAGCUAACUAGGAGAGUCCGCAAUCCUCAGUCCUCAGUUCGGAAUUUGG------- ...((((((((((....))))........(((((((((((((((..((((((.....))))))(((..(....)..)))..)))))))))))))))))))))------- ( -35.40, z-score = -1.71, R) >droSec1.super_10 1410681 98 - 3036183 GUCCCGAAUCCUUGCGCAAGGCUCGCUCU----GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCCGGAUUUUGG------- ((((.((((((..(((.......)))...----(((((((((((((..((((..(((.....)))..)))).))))))))))))).)))))).)))).....------- ( -41.00, z-score = -3.69, R) >droSim1.chrX 1137385 98 - 17042790 GUCCCGAAUCCUUGCGCAAGGCUCGCUCU----GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAUGUCCUCAGUCCGGAUUCCGGAUUUUGG------- ((((.((((((..(((.......)))...----((((((((((..((.((((..(((.....)))..)))).)).)))))))))).)))))).)))).....------- ( -38.20, z-score = -2.92, R) >consensus GUCCCGAAUCCUUGCGCAAGGCUCGCUCU____GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCCGGAUUUUGG_______ ((((.((((((..((.....))...........(((((((((((((((((((........)))))))......)))))))))))).)))))).))))............ (-28.56 = -30.28 + 1.72)

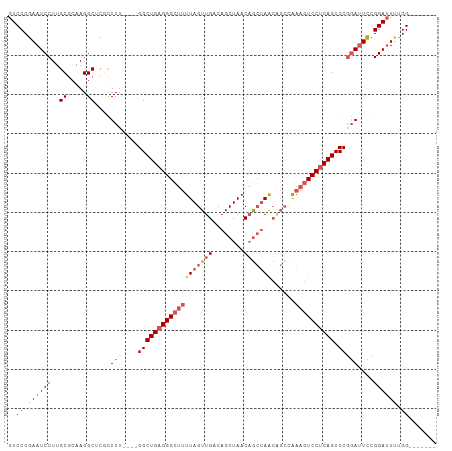
| Location | 1,606,127 – 1,606,217 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Shannon entropy | 0.25192 |
| G+C content | 0.53592 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -23.76 |
| Energy contribution | -25.28 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1606127 90 - 22422827 AGUCCCGAAUCCUUGCACAAGGCUCGCUCU----GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCC------- .....(((..((((....)))).))).(((----(((((((((((((..((((..(((.....)))..)))).))))))))))).)))))....------- ( -34.70, z-score = -3.32, R) >droEre2.scaffold_4644 1594155 97 - 2521924 AGUCCCGAAUCCUUGCGCAAGGCUCUCUCA----GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACUGGUAAAGUCCGCAGUCCGGAUUCCGGAUUCC (((((.((((((...(....).........----(((((.((((((((((((...(((.....))))))))..))))))).))))).)))))).))))).. ( -35.30, z-score = -2.09, R) >droYak2.chrX 1458796 94 - 21770863 AGUCCCGAAUCCUUGCGCAAGGCUCUUUCUUUCGGGCUGAGGGCUUUUAGUUGACAGCUAACUAGGAGAGUCCGCAAUCCUCAGUCCUCAGUUC------- ...((((((.((((....))))........))))))(((((((((..((((((.....))))))(((..(....)..)))..)))))))))...------- ( -29.30, z-score = -0.81, R) >droSec1.super_10 1410690 90 - 3036183 AGUCCCGAAUCCUUGCGCAAGGCUCGCUCU----GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCC------- ......((((((..(((.......)))...----(((((((((((((..((((..(((.....)))..)))).))))))))))))).)))))).------- ( -35.80, z-score = -3.26, R) >droSim1.chrX 1137394 90 - 17042790 AGUCCCGAAUCCUUGCGCAAGGCUCGCUCU----GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAUGUCCUCAGUCCGGAUUCC------- ......((((((..(((.......)))...----((((((((((..((.((((..(((.....)))..)))).)).)))))))))).)))))).------- ( -33.00, z-score = -2.43, R) >consensus AGUCCCGAAUCCUUGCGCAAGGCUCGCUCU____GGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCAGUCCGGAUUCC_______ ......((((((..((.....))...........(((((((((((((((((((........)))))))......)))))))))))).))))))........ (-23.76 = -25.28 + 1.52)
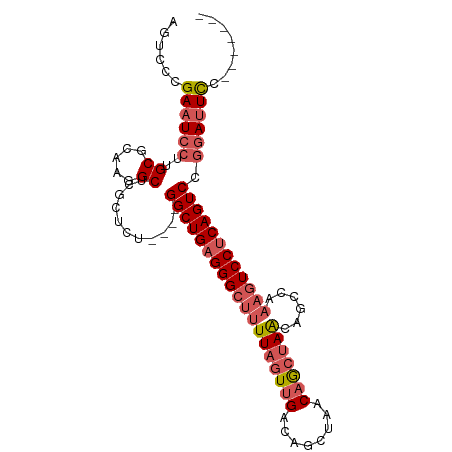
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:03 2011