
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,590,285 – 1,590,368 |
| Length | 83 |
| Max. P | 0.895976 |

| Location | 1,590,285 – 1,590,368 |
|---|---|
| Length | 83 |
| Sequences | 6 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
| Shannon entropy | 0.37597 |
| G+C content | 0.60958 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -19.49 |
| Energy contribution | -21.43 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
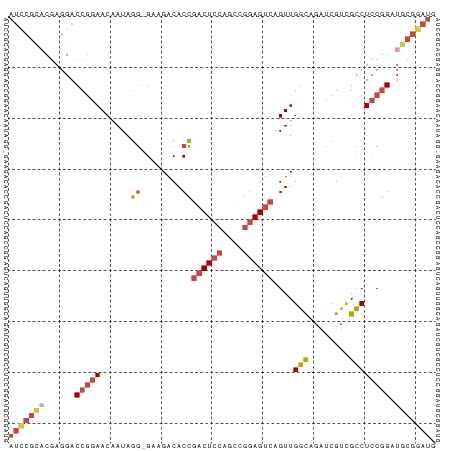
>dm3.chrX 1590285 83 + 22422827 AUCCGUACGAGGACCGGAACGAUAGG-GAAGACACCGACUCCAGCUGGAGUCAGUUGGCAGAUCGUCGCCUCCGGAUGCGGAUG (((((((......(((((.((((.((-.......))(((((((((((....))))))).)).))))))..))))).))))))). ( -32.00, z-score = -1.51, R) >droSim1.chrX_random 1054247 83 + 5698898 AUCCGCACGAGGACCGGAACGAUAGG-GAGGACACCGACUCCAGCCGGAGUCAGUUGGCAGAUCGUCGCCUCCGGAUGCGGAUG (((((((...(((..((.(((((...-(..(((...((((((....)))))).)))..)..)))))..)))))...))))))). ( -32.90, z-score = -1.23, R) >droSec1.super_10 1395665 83 + 3036183 AUCCGCACGAGGACCGGAACAAUAGG-GAGGACACCGACUCCAACCGGAGUCAGUUGGCAGAUCGUCGCCUCCGGAUGCGGAUG (((((((((.....)).........(-((((.(...((((((....))))))....(((.....)))))))))...))))))). ( -32.40, z-score = -1.77, R) >droYak2.chrX 1443588 83 + 21770863 AUUCGCCCGAGGACCGGAACAAUAGG-GAAGACACCGACUCCAGCCGGAGUCAGUUGGUAGAUCGCCGCCUCCGGAUGCGGAUG ((((((((((((..(((..((((.((-.......))((((((....)))))).))))........)))))).)))..)))))). ( -28.70, z-score = -0.65, R) >droEre2.scaffold_4644 1577178 83 + 2521924 AUUCGCACGAGGACCGGAACAAUAGG-GAAGACACCGACUCCAGCCGGAGUCAGUUGGCAGAUCGUUGCCUCCGGAUGCGGAUG (((((((......(((((......((-.......))((((((....))))))....(((((....)))))))))).))))))). ( -30.70, z-score = -1.84, R) >droAna3.scaffold_12929 1465209 72 + 3277472 ------------CCCACAACCAUAUUUGGCGGCACGAGCUCCCGCCAAAGGCAGUUGGCCAUUCGUCGUCUUCUGCCACGGAUG ------------.......((...((((((((.........))))))))(((((..(((........)))..)))))..))... ( -25.00, z-score = -2.09, R) >consensus AUCCGCACGAGGACCGGAACAAUAGG_GAAGACACCGACUCCAGCCGGAGUCAGUUGGCAGAUCGUCGCCUCCGGAUGCGGAUG (((((((......(((((..................((((((....))))))....(((........)))))))).))))))). (-19.49 = -21.43 + 1.95)


| Location | 1,590,285 – 1,590,368 |
|---|---|
| Length | 83 |
| Sequences | 6 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Shannon entropy | 0.37597 |
| G+C content | 0.60958 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -21.83 |
| Energy contribution | -23.50 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.895976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
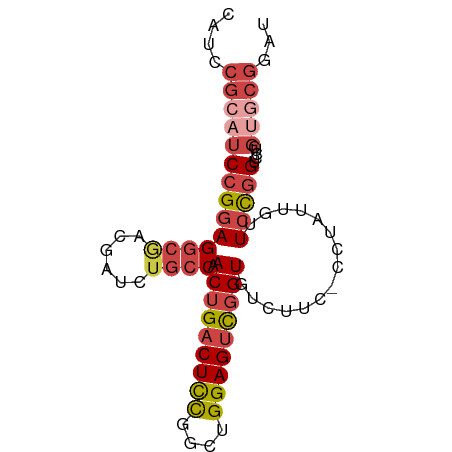
>dm3.chrX 1590285 83 - 22422827 CAUCCGCAUCCGGAGGCGACGAUCUGCCAACUGACUCCAGCUGGAGUCGGUGUCUUC-CCUAUCGUUCCGGUCCUCGUACGGAU .(((((.(((((((((((......)))).(((((((((....)))))))))......-........))))).....)).))))) ( -32.00, z-score = -1.83, R) >droSim1.chrX_random 1054247 83 - 5698898 CAUCCGCAUCCGGAGGCGACGAUCUGCCAACUGACUCCGGCUGGAGUCGGUGUCCUC-CCUAUCGUUCCGGUCCUCGUGCGGAU .(((((((((((((((((......)))).(((((((((....)))))))))......-........))))).....)))))))) ( -37.90, z-score = -2.74, R) >droSec1.super_10 1395665 83 - 3036183 CAUCCGCAUCCGGAGGCGACGAUCUGCCAACUGACUCCGGUUGGAGUCGGUGUCCUC-CCUAUUGUUCCGGUCCUCGUGCGGAU .(((((((((((((((((......)))).(((((((((....)))))))))......-........))))).....)))))))) ( -37.90, z-score = -3.13, R) >droYak2.chrX 1443588 83 - 21770863 CAUCCGCAUCCGGAGGCGGCGAUCUACCAACUGACUCCGGCUGGAGUCGGUGUCUUC-CCUAUUGUUCCGGUCCUCGGGCGAAU ....(((....((((((((.......)).(((((((((....)))))))))))))))-.........(((.....))))))... ( -28.70, z-score = -0.03, R) >droEre2.scaffold_4644 1577178 83 - 2521924 CAUCCGCAUCCGGAGGCAACGAUCUGCCAACUGACUCCGGCUGGAGUCGGUGUCUUC-CCUAUUGUUCCGGUCCUCGUGCGAAU ....((((((((((((((......)))).(((((((((....)))))))))......-........))))).....)))))... ( -33.40, z-score = -2.87, R) >droAna3.scaffold_12929 1465209 72 - 3277472 CAUCCGUGGCAGAAGACGACGAAUGGCCAACUGCCUUUGGCGGGAGCUCGUGCCGCCAAAUAUGGUUGUGGG------------ (((.(((.((....).).))).))).(((...(((((((((((.(.....).))))))))...)))..))).------------ ( -23.50, z-score = -0.14, R) >consensus CAUCCGCAUCCGGAGGCGACGAUCUGCCAACUGACUCCGGCUGGAGUCGGUGUCUUC_CCUAUUGUUCCGGUCCUCGUGCGGAU ....((((((((((((((......)))).(((((((((....)))))))))...............))))).....)))))... (-21.83 = -23.50 + 1.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:57 2011