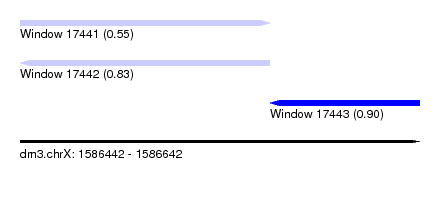
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,586,442 – 1,586,642 |
| Length | 200 |
| Max. P | 0.901186 |

| Location | 1,586,442 – 1,586,567 |
|---|---|
| Length | 125 |
| Sequences | 5 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Shannon entropy | 0.15110 |
| G+C content | 0.51101 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -30.75 |
| Energy contribution | -31.11 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
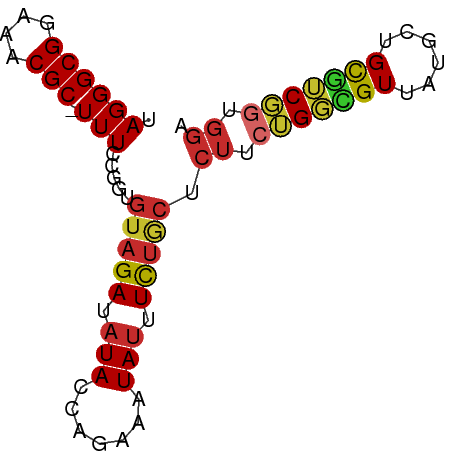
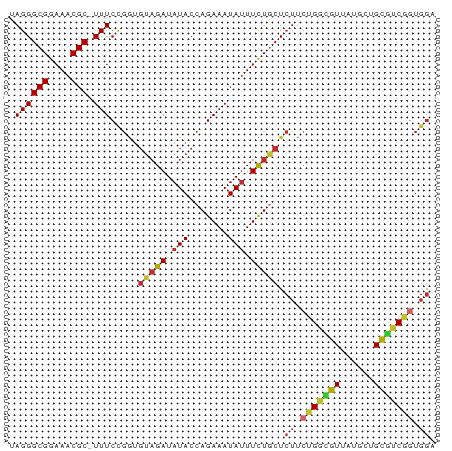
>dm3.chrX 1586442 125 + 22422827 UCUAGGUGCUCAUAGCUCGAGGAGCUCACCGAUUUAGCCGUGUCCUUAGGAUAAAGAUUCGAUUCAAUUCGCUUCGCUUUGCACUCGUCACUACACAGCUUUCUGGAG-------UCAGGACACGGCACUCG ....((((.....(((((...)))))))))......((((((((((..((((....))))(((((.....((........)).............(((....))))))-------))))))))))))..... ( -39.30, z-score = -2.03, R) >droEre2.scaffold_4644 1573342 127 + 2521924 UCUAGGCGCUCAUAGCUCGAGGAGCUCACCGAUUUAGCCGUGUCCUUAGGAUUUCGAUUCGAUUCAAUUCGCUUCGC-----ACUCGUCACUACACAGCUUUCAGGAGUCAGGAGUCAGGACACGGCACUCG ....((.(.....(((((...)))))).))......((((((((((...((((((((..(((......)))..))(.-----((((...................))))).))))))))))))))))..... ( -37.31, z-score = -1.13, R) >droYak2.chrX 1439703 120 + 21770863 UCUAGGUGCUCAUAGCUCGACGAGCUCACCGAUUUAGCCGUGUCCUUAGGAUUAAGAUUCGAUUCAAUUCGCUUCGC-----ACUCGUCACUACACAGCUUUCAGGAG-------UCAGGACACGGCACUCG ....((((.....(((((...)))))))))......((((((((((((((((...((..(((......)))..))..-----....))).)))....((((....)))-------).))))))))))..... ( -36.20, z-score = -2.19, R) >droSec1.super_10 1391844 132 + 3036183 UCUAGGUGCUCAUAGCUCGAGGAGCUCACCGAUUUAGCCGUGUCCUUAGGAUCUAGAUUCAAUUCAAUUCGCUCCGCUUUGCACUCGUCACUACACAGCUUUCUGGAGUCAGGAGUCAGGACACGGCACUCG ....((((.....(((((...)))))))))......((((((((((..((((....))))..........(((((((...))((((((....)).(((....)))))))..))))).))))))))))..... ( -41.70, z-score = -1.76, R) >droSim1.chrX_random 1050400 132 + 5698898 UCUAGGUGCUCAUAGCUCGAGGAGCUCACCGAUUUAGCCGUGUCCUUAGGAUAUAGAUUCAAUUAAAUUCGCUCCGCUUUGCACUCGUCACUACACAGCUUUCUGGAGUCAGGAGUCAGGACACGGCACUCG ....((((.....(((((...)))))))))......((((((((((..((((....))))..........(((((((...))((((((....)).(((....)))))))..))))).))))))))))..... ( -41.10, z-score = -1.86, R) >consensus UCUAGGUGCUCAUAGCUCGAGGAGCUCACCGAUUUAGCCGUGUCCUUAGGAUAUAGAUUCGAUUCAAUUCGCUUCGCUUUGCACUCGUCACUACACAGCUUUCUGGAGUCAGGAGUCAGGACACGGCACUCG ....((((.....(((((...)))))))))......((((((((((..((((....))))..........(((((.............................)))))........))))))))))..... (-30.75 = -31.11 + 0.36)
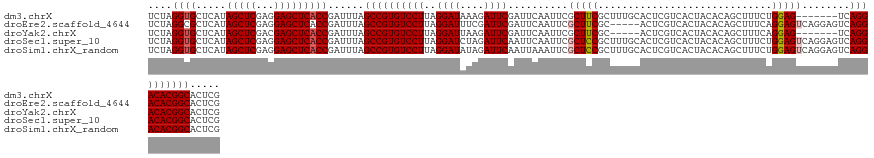
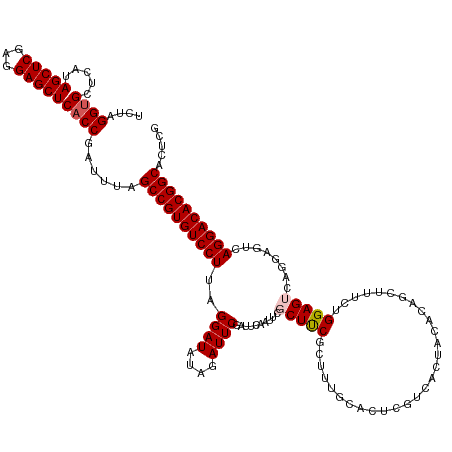
| Location | 1,586,442 – 1,586,567 |
|---|---|
| Length | 125 |
| Sequences | 5 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Shannon entropy | 0.15110 |
| G+C content | 0.51101 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -35.18 |
| Energy contribution | -36.14 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1586442 125 - 22422827 CGAGUGCCGUGUCCUGA-------CUCCAGAAAGCUGUGUAGUGACGAGUGCAAAGCGAAGCGAAUUGAAUCGAAUCUUUAUCCUAAGGACACGGCUAAAUCGGUGAGCUCCUCGAGCUAUGAGCACCUAGA .((..((((((((((..-------.........(((.((((.(....).)))).)))((..(((......)))..)).........))))))))))....))(((((((((...))))).....)))).... ( -40.40, z-score = -2.02, R) >droEre2.scaffold_4644 1573342 127 - 2521924 CGAGUGCCGUGUCCUGACUCCUGACUCCUGAAAGCUGUGUAGUGACGAGU-----GCGAAGCGAAUUGAAUCGAAUCGAAAUCCUAAGGACACGGCUAAAUCGGUGAGCUCCUCGAGCUAUGAGCGCCUAGA .((..((((((((((.((((.(.(((((........).).))).).))))-----.(((..(((......)))..)))........))))))))))....))(((((((((...))))).....)))).... ( -40.80, z-score = -2.06, R) >droYak2.chrX 1439703 120 - 21770863 CGAGUGCCGUGUCCUGA-------CUCCUGAAAGCUGUGUAGUGACGAGU-----GCGAAGCGAAUUGAAUCGAAUCUUAAUCCUAAGGACACGGCUAAAUCGGUGAGCUCGUCGAGCUAUGAGCACCUAGA .((..((((((((((.(-------(((......(((....)))...))))-----..((..(((......)))..)).........))))))))))....))(((..((((((......))))))))).... ( -36.70, z-score = -1.34, R) >droSec1.super_10 1391844 132 - 3036183 CGAGUGCCGUGUCCUGACUCCUGACUCCAGAAAGCUGUGUAGUGACGAGUGCAAAGCGGAGCGAAUUGAAUUGAAUCUAGAUCCUAAGGACACGGCUAAAUCGGUGAGCUCCUCGAGCUAUGAGCACCUAGA .((..((((((((((.((((.(.(((((((....))).).))).).)))).......(((..((.((.....)).))....)))..))))))))))....))(((((((((...))))).....)))).... ( -41.30, z-score = -1.38, R) >droSim1.chrX_random 1050400 132 - 5698898 CGAGUGCCGUGUCCUGACUCCUGACUCCAGAAAGCUGUGUAGUGACGAGUGCAAAGCGGAGCGAAUUUAAUUGAAUCUAUAUCCUAAGGACACGGCUAAAUCGGUGAGCUCCUCGAGCUAUGAGCACCUAGA .((..((((((((((.((((.(.(((((((....))).).))).).)))).......(((....((((....)))).....)))..))))))))))....))(((((((((...))))).....)))).... ( -41.80, z-score = -1.85, R) >consensus CGAGUGCCGUGUCCUGACUCCUGACUCCAGAAAGCUGUGUAGUGACGAGUGCAAAGCGAAGCGAAUUGAAUCGAAUCUAAAUCCUAAGGACACGGCUAAAUCGGUGAGCUCCUCGAGCUAUGAGCACCUAGA .((..((((((((((..................(((.((((.(....).)))).)))((..(((......)))..)).........))))))))))....))(((((((((...))))).....)))).... (-35.18 = -36.14 + 0.96)
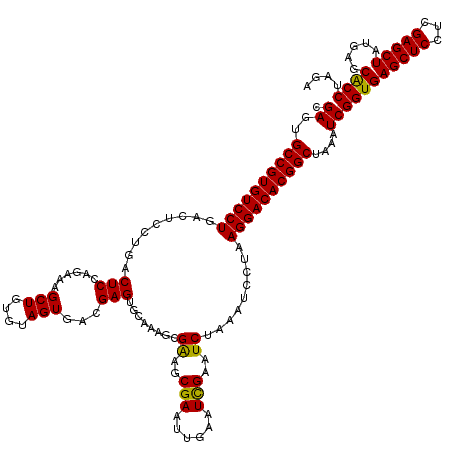
| Location | 1,586,567 – 1,586,642 |
|---|---|
| Length | 75 |
| Sequences | 9 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 85.69 |
| Shannon entropy | 0.29067 |
| G+C content | 0.46603 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -15.24 |
| Energy contribution | -14.81 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1586567 75 - 22422827 UAGGGCGGAAACGC-UUUCCGGUGUAGAUAUACCAGAAAUAUUUCUGCUCUUCUGGCGUUAUGCUGCGUCGGUGGA .((((((((((...-((((.(((((....))))).))))..)))))))))).(((((((......))))))).... ( -28.00, z-score = -2.87, R) >droSim1.chrX_random 1050532 75 - 5698898 UAGGGCGGAAACGC-UUUCCGGUGUAGAUAUACCAGAAAUAUUUCUGCUCUUCUGGCGUUAUGCUGCGUCGGUGGA .((((((((((...-((((.(((((....))))).))))..)))))))))).(((((((......))))))).... ( -28.00, z-score = -2.87, R) >droSec1.super_10 1391976 75 - 3036183 UAGGGCGGAAACGC-UUUCCGGUGUAGAUAUACCAGAAAUAUUUCUGCUCUUCUGGCGUUAUGCUGCGUCGGUGGA .((((((((((...-((((.(((((....))))).))))..)))))))))).(((((((......))))))).... ( -28.00, z-score = -2.87, R) >droYak2.chrX 1439823 75 - 21770863 UAGGGCGGAAACGC-UUUCCGGUGUAGAUAUACCAGAAAUAUUUCUGCUCUUCUGGCGUUAUGCUGCAUCGGUGGA .((((((....)))-)))((((((((((((..((((((............))))))...))).))))))))).... ( -28.90, z-score = -3.45, R) >droEre2.scaffold_4644 1573469 75 - 2521924 UAGGGCGGAAACGC-UUUCCGGUGUAGAUAUACCAGAAAUAUUUCUGCUCUUCUGGCGUUAUGCUGCGUCGGUGGA .((((((((((...-((((.(((((....))))).))))..)))))))))).(((((((......))))))).... ( -28.00, z-score = -2.87, R) >droAna3.scaffold_12929 1459187 75 - 3277472 UAGGGCGGAAACGC-UUUCCGGUGUAGAUAUACCAGAAAUAUUUUUGCUCUCCAGCCGUUCUGCUGCGGCUCUGGA ..(((((((((...-((((.(((((....))))).))))..)))))))))(((((((((......)))))..)))) ( -27.40, z-score = -2.35, R) >dp4.chrXL_group1a 4754510 75 + 9151740 UAGGGCGGAAACGCUUUUCUGAUGUAGAUAUACCAGAAAUAUUUUUACUCUUCUGCUGUUCUACUGUGGCGGUGG- ...((((....))))((((((.(((....))).))))))..........((.((((..(......)..)))).))- ( -19.10, z-score = -1.60, R) >droPer1.super_11 1870004 75 + 2846995 UAGGGCGGAAACGCUUUUCUGAUGUAGAUAUACCAGAAAUAUUUUUACUCUUCUGCUGUUCUACUGUGGCGGUGG- ...((((....))))((((((.(((....))).))))))..........((.((((..(......)..)))).))- ( -19.10, z-score = -1.60, R) >droGri2.scaffold_14853 680681 63 - 10151454 UAGGGCGGAAACGC-UUUCU------GAUAUACCAGAAAUAAUUUUGCUGUUGAG-UGUUAAAACGUAUCG----- .((((((....)))-)))..------(((((..((((((....))).)))....(-(......))))))).----- ( -13.50, z-score = -1.38, R) >consensus UAGGGCGGAAACGC_UUUCCGGUGUAGAUAUACCAGAAAUAUUUCUGCUCUUCUGGCGUUAUGCUGCGUCGGUGGA ..(((((....)))....))...(((((.(((.......))).))))).((.(((((((......))))))).)). (-15.24 = -14.81 + -0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:55 2011