
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,584,019 – 1,584,128 |
| Length | 109 |
| Max. P | 0.968518 |

| Location | 1,584,019 – 1,584,128 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 60.12 |
| Shannon entropy | 0.81629 |
| G+C content | 0.48724 |
| Mean single sequence MFE | -35.69 |
| Consensus MFE | -12.83 |
| Energy contribution | -13.35 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
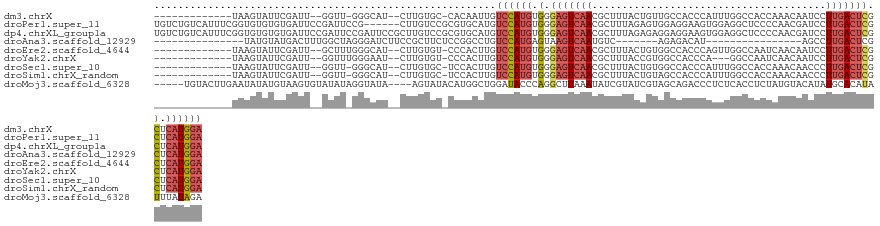
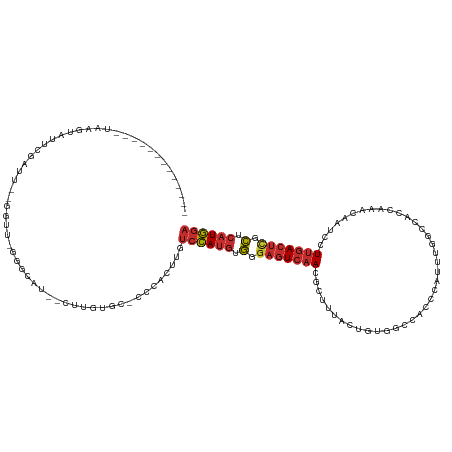
>dm3.chrX 1584019 109 + 22422827 -------------UAAGUAUUCGAUU--GGUU-GGGCAU--CUUGUGC-CACAAUUGUCCAUGUGGGAGUCAACGCUUUACUGUUGCCACCCAUUUGGCCACCAAACAAUCCUUGACUCGCUCAUGGA -------------........(((((--(...-.(((((--...))))-).))))))((((((.(.(((((((.........((.((((......)))).))..........))))))).).)))))) ( -35.51, z-score = -2.13, R) >droPer1.super_11 1867040 122 - 2846995 UGUCUGUCAUUUCGGUGUGUGUGAUUCCGAUUCCG------CUUGUCCGCGUGCAUGUCCAUGUGGGAGUCAACGCUUUAGAGUGGAGGAAGUGGAGGCUCCCCAACGAUCCUUGACUCGCUCAUGGA ..............(..((((.(((..((....))------...)))))))..)...((((((.(.(((((..((((....))))(((((..(((.(....))))....)))))))))).).)))))) ( -40.00, z-score = -0.21, R) >dp4.chrXL_group1a 4751557 128 - 9151740 UGUCUGUCAUUUCGGUGUGUGUGAUUCCGAUUCCGAUUCCGCUUGUCCGCGUGCAUGUCCAUGUGGGAGUCAACGCUUUAGAGAGGAGGAAGUGGAGGCUCCCCAACGAUCCUUGACUCGCUCAUGGA .((..((((..((((..((((.(((..((..........))...)))))))..).......((.(((((((..((((((.........))))))..))))))))).)))....))))..))....... ( -37.10, z-score = 1.18, R) >droAna3.scaffold_12929 1456332 90 + 3277472 ---------------UAUGUAUGACUUUGGCUAGGGAUCUUCCGCUUCUCCGGCCUGUCCAUGAGUAAGUCAAUGUC-------AGAGACAU----------------AGCCUUGACUCGCUCAUGGA ---------------.............(((..((((.........))))..)))..((((((((..((((((((((-------...)))))----------------.....)))))..)))))))) ( -32.70, z-score = -2.69, R) >droEre2.scaffold_4644 1570933 110 + 2521924 -------------UAAGUAUUCGAUU--GCUUUGGGCAU--CUUGUGU-CCCACUUGUCCAUGUGGGAGUCAACGCUUUACUGUGGCCACCCAGUUGGCCAAUCAACAAUCCUUGACUCGCUCAUGGA -------------.(((((......)--))))(((((((--...))).-))))....((((((.(.(((((((..........((((((......))))))...........))))))).).)))))) ( -37.50, z-score = -2.56, R) >droYak2.chrX 1437242 107 + 21770863 -------------UAAGUAUUCGAUU--GGUUUGGGAAU--CUUGUGU-CCCACUUGUCCAUGUGGGAGUCAACGCUUUACCGUGGCCACCCA---GGCCAAUCAACAAUCCUUGACUCGCUCAUGGA -------------.........((((--((((((((...--......(-(((((........))))))((((.((......))))))..))))---)))))))).....(((.(((.....))).))) ( -38.00, z-score = -2.71, R) >droSec1.super_10 1389410 109 + 3036183 -------------UAAGUAUUCGAUU--GGUU-GGGCAU--CUUGUGC-UCCACUUGUCCAUGUGGGAGUCAACGCUUUACUGUGGCCACCCAUUUGGCCACCAAACAACCCUUGACUCGCUCAUGGA -------------.............--(((.-((((((--...))))-)).)))..((((((.(.(((((((.........(((((((......)))))))..........))))))).).)))))) ( -38.91, z-score = -2.66, R) >droSim1.chrX_random 1047215 109 + 5698898 -------------UAAGUAUUCGAUU--GGUU-GGGCAU--CUUGUGC-UCCACUUGUCCAUGUGGGAGUCAACGCUUUACUGUAGCCACCCAUUUGGCCACCAAACAACCCUUGACUCGCUCAUGGA -------------.............--(((.-((((((--...))))-)).)))..((((((.(.(((((((.........((.((((......)))).))..........))))))).).)))))) ( -32.41, z-score = -1.17, R) >droMoj3.scaffold_6328 4287599 119 + 4453435 -----UGUACUUGAAUAUAUGUAAGUGUAUAUAGGUAUA----AGUAUACAUGGCUGGAUACCCAGGCUUAAAUAUCGUAUCGUAGCAGACCCUCUCACCUCUAUGUACAUAUGCACAUAUUUAUAGA -----....(((((((((.((((.((((((((((((((.----...))))...(((((((((...............))))).))))..............)))))))))).)))).))))))).)). ( -29.06, z-score = -0.60, R) >consensus _____________UAAGUAUUCGAUU__GGUU_GGGCAU__CUUGUGC_CCCACUUGUCCAUGUGGGAGUCAACGCUUUACUGUGGCCACCCAUUUGGCCACCAAACAAUCCUUGACUCGCUCAUGGA .........................................................((((((.(.(((((((.......................................))))))).).)))))) (-12.83 = -13.35 + 0.52)
| Location | 1,584,019 – 1,584,128 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 60.12 |
| Shannon entropy | 0.81629 |
| G+C content | 0.48724 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -13.83 |
| Energy contribution | -13.33 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
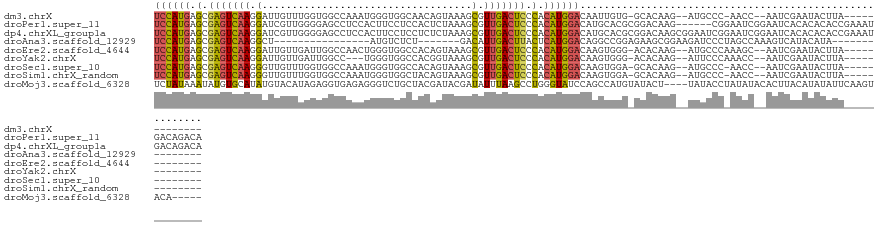
>dm3.chrX 1584019 109 - 22422827 UCCAUGAGCGAGUCAAGGAUUGUUUGGUGGCCAAAUGGGUGGCAACAGUAAAGCGUUGACUCCCACAUGGACAAUUGUG-GCACAAG--AUGCCC-AACC--AAUCGAAUACUUA------------- ((((((.(.(((((((.(.....(((...((((......))))..))).....).))))))).).))))))...(((.(-(((....--.)))))-))..--.............------------- ( -34.20, z-score = -1.39, R) >droPer1.super_11 1867040 122 + 2846995 UCCAUGAGCGAGUCAAGGAUCGUUGGGGAGCCUCCACUUCCUCCACUCUAAAGCGUUGACUCCCACAUGGACAUGCACGCGGACAAG------CGGAAUCGGAAUCACACACACCGAAAUGACAGACA ((((((.(.(((((((.(...((.((((((.......)))))).)).......).))))))).).))))))(((...(((......)------))...((((...........)))).)))....... ( -33.40, z-score = -0.35, R) >dp4.chrXL_group1a 4751557 128 + 9151740 UCCAUGAGCGAGUCAAGGAUCGUUGGGGAGCCUCCACUUCCUCCUCUCUAAAGCGUUGACUCCCACAUGGACAUGCACGCGGACAAGCGGAAUCGGAAUCGGAAUCACACACACCGAAAUGACAGACA ((((((.(.(((((((.(....((((((((.............))))))))..).))))))).).))))))......(((......)))...(((...((((...........))))..)))...... ( -36.22, z-score = -0.25, R) >droAna3.scaffold_12929 1456332 90 - 3277472 UCCAUGAGCGAGUCAAGGCU----------------AUGUCUCU-------GACAUUGACUUACUCAUGGACAGGCCGGAGAAGCGGAAGAUCCCUAGCCAAAGUCAUACAUA--------------- ((((((((..(((((.....----------------(((((...-------))))))))))..))))))))..(((.(((....(....).)))...))).............--------------- ( -31.70, z-score = -3.03, R) >droEre2.scaffold_4644 1570933 110 - 2521924 UCCAUGAGCGAGUCAAGGAUUGUUGAUUGGCCAACUGGGUGGCCACAGUAAAGCGUUGACUCCCACAUGGACAAGUGGG-ACACAAG--AUGCCCAAAGC--AAUCGAAUACUUA------------- ((((((.(.(((((((.(....(((..((((((......))))))...)))..).))))))).).))))))....((((-.((....--.))))))....--.............------------- ( -35.50, z-score = -1.87, R) >droYak2.chrX 1437242 107 - 21770863 UCCAUGAGCGAGUCAAGGAUUGUUGAUUGGCC---UGGGUGGCCACGGUAAAGCGUUGACUCCCACAUGGACAAGUGGG-ACACAAG--AUUCCCAAACC--AAUCGAAUACUUA------------- ((((((.(.(((((((.(.(((..(..(((((---.....))))))..)))..).))))))).).))))))....((((-(......--..)))))....--.............------------- ( -35.70, z-score = -1.87, R) >droSec1.super_10 1389410 109 - 3036183 UCCAUGAGCGAGUCAAGGGUUGUUUGGUGGCCAAAUGGGUGGCCACAGUAAAGCGUUGACUCCCACAUGGACAAGUGGA-GCACAAG--AUGCCC-AACC--AAUCGAAUACUUA------------- ((((((.(.(((((((.(.((.....(((((((......)))))))....)).).))))))).).))))))....(((.-(((....--.)))))-)...--.............------------- ( -40.40, z-score = -2.70, R) >droSim1.chrX_random 1047215 109 - 5698898 UCCAUGAGCGAGUCAAGGGUUGUUUGGUGGCCAAAUGGGUGGCUACAGUAAAGCGUUGACUCCCACAUGGACAAGUGGA-GCACAAG--AUGCCC-AACC--AAUCGAAUACUUA------------- ((((((.(.(((((((.(.((.....(((((((......)))))))....)).).))))))).).))))))....(((.-(((....--.)))))-)...--.............------------- ( -38.10, z-score = -2.20, R) >droMoj3.scaffold_6328 4287599 119 - 4453435 UCUAUAAAUAUGUGCAUAUGUACAUAGAGGUGAGAGGGUCUGCUACGAUACGAUAUUUAAGCCUGGGUAUCCAGCCAUGUAUACU----UAUACCUAUAUACACUUACAUAUAUUCAAGUACA----- ......((((((((.(..((((.((((..(((((.(..((......))..).........(.((((....)))).).......))----)))..)))).))))..).))))))))........----- ( -22.10, z-score = 1.31, R) >consensus UCCAUGAGCGAGUCAAGGAUUGUUGGGUGGCCAAAUGGGUGGCCACAGUAAAGCGUUGACUCCCACAUGGACAAGUGGG_GCACAAG__AUGCCC_AACC__AAUCGAAUACUUA_____________ ((((((.(.(((((((.(...................................).))))))).).))))))......................................................... (-13.83 = -13.33 + -0.50)
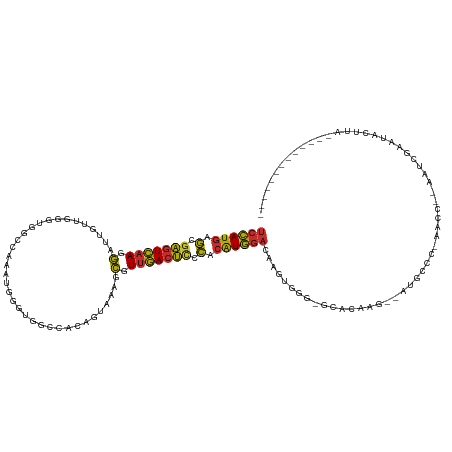
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:52 2011