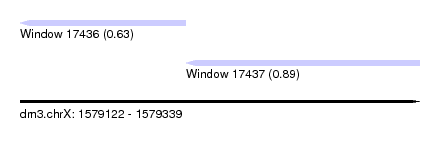
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,579,122 – 1,579,339 |
| Length | 217 |
| Max. P | 0.890437 |

| Location | 1,579,122 – 1,579,212 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.87 |
| Shannon entropy | 0.16548 |
| G+C content | 0.41489 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -17.28 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1579122 90 - 22422827 AGAAUUUGCCUUUCUACCCACUUAUAGCUAGCAUUGUACGAUCGCUUCCAUUU------------ACCCAAAGUGUGUGUGCAAUAUAAGCGGUGACAUUUC ((((.......))))...((((....(((.(.((((((((..(((((......------------.....)))))..)))))))).).)))))))....... ( -16.60, z-score = -0.55, R) >droEre2.scaffold_4644 1564553 90 - 2521924 AGAAUUUGCCUUUCUAUCCACUUGUAGCUAGCAUUGUACGAUCGCUUCCAUGU------------ACCCAAUGUAUGUGUGCAAUGUAAGCAGUGACAUUUC ((((.......))))...((((....(((.(((((((((((.....)).((((------------((.....))))))))))))))).)))))))....... ( -20.50, z-score = -1.42, R) >droYak2.chrX 1432220 100 - 21770863 AGAAUUUGCCUUUCUACCCACUUAUAGCUAGCAU--UACGAUCGCUUCCAUGUACCCACCCAAUGACCCAAAGUAUGUGUGCAAUGUAAGCAGUGACAUUUC ((((.......))))...((((....(((.((((--(......((...(((((((.................))))))).))))))).)))))))....... ( -13.43, z-score = 0.37, R) >droSec1.super_10 1384535 90 - 3036183 AGAAUUUGCCUUUCUACCCACUUAUAGCUAGCAUUGUACGAUCGCUUCCAUUU------------AACCAAAGUGUGUGUGCAAUGUAAGCGGUGACAUUUC ((((.......))))...((((....(((.((((((((((..(((((......------------.....)))))..)))))))))).)))))))....... ( -22.90, z-score = -2.35, R) >droSim1.chrX_random 1041833 90 - 5698898 AGAAUUUGCCUUUCUACCCACUUAUAGCUAGCAUUGUACGAUCGCUUCCAUUU------------AACCAAAGUGUGUGUGCAAUGUAAGCGGUGACAUUUC ((((.......))))...((((....(((.((((((((((..(((((......------------.....)))))..)))))))))).)))))))....... ( -22.90, z-score = -2.35, R) >consensus AGAAUUUGCCUUUCUACCCACUUAUAGCUAGCAUUGUACGAUCGCUUCCAUUU____________ACCCAAAGUGUGUGUGCAAUGUAAGCGGUGACAUUUC ((((.......))))...((((....(((.((((((((((..(((((.......................)))))..)))))))))).)))))))....... (-17.28 = -18.24 + 0.96)

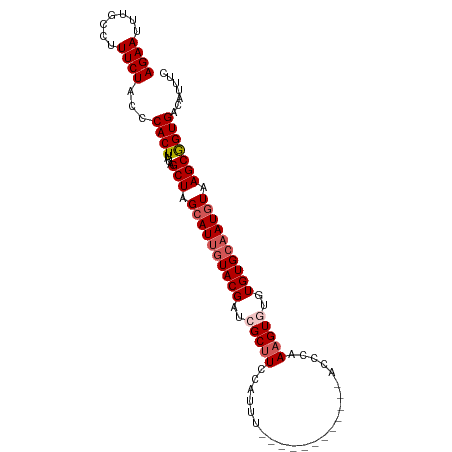

| Location | 1,579,212 – 1,579,339 |
|---|---|
| Length | 127 |
| Sequences | 9 |
| Columns | 129 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Shannon entropy | 0.49213 |
| G+C content | 0.49346 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.36 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1579212 127 - 22422827 UGUAUCUGAACCAACGAACCGAAAAAGGAGCGCCUGUGAAACCACUCAGAUCGCUUAAUCCAAGGGGUAGCAUGGAACCCCUGUUCCCCUACACCCCGGAUCAGUUAUACAGCAUUCCAGGUACACA-- ((((((((.......((.(((......(((((.(((.(......).)))..))))).......(((((((...(((((....))))).)))).)))))).)).(((....)))....))))))))..-- ( -36.80, z-score = -2.22, R) >droWil1.scaffold_181096 2876248 116 + 12416693 --------UAUAUAUGAA---AAAAAGGAGCGCCUGUGAAACCUCUUAGAUCGCUUAAUAGCAGGGGUAGCAUGGAACCCCUGUUUCCCUAUACCCCGGAUCAGUUAUACAGUAUUCCAGGUACACA-- --------..........---........(..((((.(((........(((((((....))).((((((....(((((....)))))....)))))).)))).(.....)....)))))))..)...-- ( -30.40, z-score = -1.16, R) >droPer1.super_11 1861738 125 + 2846995 --CACCCCAAAAACAUACCCUGUAAAGGAGCGCCUGUGAAACCACUAAGAUCAGUCAAUCCAAGGGGUAGCAUGGAACCCCUGUUUCCCUAUACCCAGGAUCAGUUAUACAGCAUUCCAGGUACACC-- --.(((......(((.....)))...((((.((.((((.(((......((....)).((((...(((((....(((((....)))))....))))).))))..))).)))))).)))).))).....-- ( -27.50, z-score = -0.25, R) >dp4.chrXL_group1a 4746370 125 + 9151740 --CACCCCAAAAACAUACCCUGUAAAGGAGCGCCUGUGAAACCACUAAGAUCAGUCAAUCCAAGGGGUAGCAUGGAACCCCUGUUUCCCUAUACCCAGGAUCAGUUAUACAGCAUUCCAGGUACACC-- --.(((......(((.....)))...((((.((.((((.(((......((....)).((((...(((((....(((((....)))))....))))).))))..))).)))))).)))).))).....-- ( -27.50, z-score = -0.25, R) >droAna3.scaffold_12929 1450436 128 - 3277472 -UUUAUCAAAAUCGAAAAUAUAAAAAGGAGCGCCAGUGAAACCACUCAGAUCACUCAAUUCAAGGGGUAGCAUGGAACCCCUGUUCCCCUACACCCCGGAUCAGUUAUACAGCAUUCCAGGUACACACG -.........................((((.((.((((....))))..((((..........(((((.((((.((....)))))))))))........)))).........)).))))........... ( -26.47, z-score = -0.96, R) >droEre2.scaffold_4644 1564643 125 - 2521924 --UAUCUGAACCAACGAACCGAAAAAGGAGCGCCUGUGAAACCACUCAGAUCACUUAAUCCAAGGGGUAGCAUGGAACCCCUGUUCCCCUACACCCCGGAUCAGUUAUACAGCAUUCCAGGUACACG-- --((((((.......((.(((.....(((......((((...........))))....)))..(((((((...(((((....))))).)))).)))))).)).(((....)))....))))))....-- ( -30.30, z-score = -1.13, R) >droSec1.super_10 1384625 127 - 3036183 UGUAUCUGAACCAACGAACCGAAAAAGGAGCGCCUGUGAAACCACUCAGAUCGCUUAAUCCAAGGGGUAGCAUGGAACCCCUGUUCCCCUACACCCCGGAUCAGUUAUACAGCAUUCCAGGUACACG-- ((((((((.......((.(((......(((((.(((.(......).)))..))))).......(((((((...(((((....))))).)))).)))))).)).(((....)))....))))))))..-- ( -36.80, z-score = -2.34, R) >droSim1.chrX_random 1041923 127 - 5698898 UGUAUCUGAACCAACGAACCGAAAAAGGAGCGCCUGUGAAACCACUCAGAUCGCUUAAUCCAAGGGGUAGCAUGGAACCCCUGUUCCCCUACACCCCGGAUCAGUUAUACAGCAUUCCAGGUACACG-- ((((((((.......((.(((......(((((.(((.(......).)))..))))).......(((((((...(((((....))))).)))).)))))).)).(((....)))....))))))))..-- ( -36.80, z-score = -2.34, R) >anoGam1.chr2L 8116247 98 - 48795086 -------------------------AGGAGUAGCAUCGAAACCGAUCCGCUCGUACGGUUCACGCGGCAACAUUGCCGGCCCGAUGCCAUACACUACCGAGCAAUUGUACAACAUUCCAGGUA------ -------------------------.((((((((((((....))))..))).((((((((...((((((....))))(((.....)))............))))))))))...))))).....------ ( -27.60, z-score = -0.93, R) >consensus __UAUCUGAACCAACGAACCGAAAAAGGAGCGCCUGUGAAACCACUCAGAUCGCUUAAUCCAAGGGGUAGCAUGGAACCCCUGUUCCCCUACACCCCGGAUCAGUUAUACAGCAUUCCAGGUACACA__ ..........................((((.((..(((....)))...((((..........(((((.((((.((....)))))))))))........)))).........)).))))........... (-20.08 = -20.36 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:50 2011