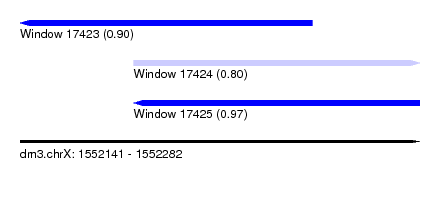
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,552,141 – 1,552,282 |
| Length | 141 |
| Max. P | 0.968119 |

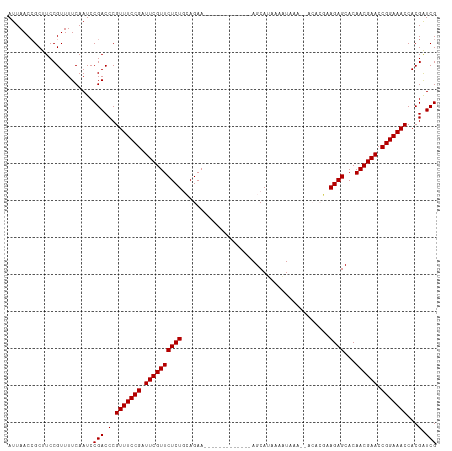
| Location | 1,552,141 – 1,552,244 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.87 |
| Shannon entropy | 0.56558 |
| G+C content | 0.40299 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1552141 103 - 22422827 -------------AUUUUAUGCUUUCUGAAGAGAACGAACCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAUUUUGUCGAUGGCUUGAUACGUUUGAGCCUAGAUUUUGACG -------------......(((((((.........(((.(((....))).)))..........)))))))(((((....(((...((((..(.....)..))))..))).))))). ( -32.11, z-score = -3.14, R) >droPer1.super_11 1827122 81 + 2846995 ------------------AUGAUUCC---AGCUAAGUAAUCGCGAGAGAAUAUAGUACAGAACGGAAACGGUU-------UUGGGGGGUAAUUAAUUUUGAAAC--GUUUU----- ------------------...(((((---.((((.((..((....))..)).)))).(((((((....).)))-------))).)))))...............--.....----- ( -17.80, z-score = -1.80, R) >dp4.chrXL_group1a 4714068 81 + 9151740 ------------------AUGAUUCC---AGCUAAGUAAUCGCGAGAGAAUAUAGUACAGAACGGAAACGGUU-------UUGGGGGGUAAUUAAUUUUGAAAC--GUUUU----- ------------------...(((((---.((((.((..((....))..)).)))).(((((((....).)))-------))).)))))...............--.....----- ( -17.80, z-score = -1.80, R) >droEre2.scaffold_4644 1537345 102 - 2521924 -------------AUUAUAUGCUUUCUUCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAUUUUGUCGUAGGCUUGAUAUGUUUGAACCUCG-UUUUGACG -------------......((((((((((((....(((..((....))..)))..)))))...)))))))(((((......((.(((.(..(.....)..).)))))-..))))). ( -25.20, z-score = -0.63, R) >droYak2.chrX 1405369 116 - 21770863 AAUUUCUGCUUUAAUGCAGAAUAUUCUCCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCUGUUAAUUUUGUCGAUGGCUUGAUAUGUUUGAACCUCGAUUUUGACG ...((((((......))))))..((((....))))(((..((....))..))).........(....)..(((((....(((((.((.(..(.....)..).))))))).))))). ( -29.80, z-score = -1.40, R) >droSec1.super_10 1357830 103 - 3036183 -------------AUUUUAUGCUUUCUGCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAUUUUGUCGAUGGCUUGAUACGUUUGAACCUCGAUUUUAACG -------------......(((((((..(((....(((..((....))..)))..))).....)))))))(((((....(((((.((.(..(.....)..).))))))).))))). ( -27.00, z-score = -1.22, R) >droSim1.chrX_random 1021331 103 - 5698898 -------------AUUUUCUGCUUUCUGCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAUUUUGUCGAUGGCUUGAUACGUUUGAACCUCGAUUUUAACG -------------.((((((((.....))))))))(((..((....))..))).........((....))(((((....(((((.((.(..(.....)..).))))))).))))). ( -31.10, z-score = -2.28, R) >consensus _____________AUUUUAUGCUUUCUGCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAUUUUGUCGAUGGCUUGAUACGUUUGAACCUCGAUUUUGACG ....................((((((......((.(((.(((....))).)))..))......))))))((((.....((((((....))))))......))))............ (-11.65 = -11.70 + 0.05)

| Location | 1,552,181 – 1,552,282 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.88 |
| Shannon entropy | 0.18222 |
| G+C content | 0.44488 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1552181 101 + 22422827 AUUAACCGCUUCCGUUUUCAAUCCGACCCGUUUCCGGUUCGUUCUCUUCAGAA-------------AGCAUAAAAUAAA--ACACGAAGAGCACAACGAACCGGAAACCACGAUCG .......................(((.(.((((((((((((((((((((....-------------.............--....))))))...))))))))))))))...).))) ( -28.10, z-score = -4.23, R) >droEre2.scaffold_4644 1537384 101 + 2521924 AUUAACCGCUUCCGUUUUCAAUCCGACCCGUUUCCGAUUCGUUCUCUGAAGAA-------------AGCAUAUAAUAAA--GAAAUAAGAGCACAACGAACCGGAAACCACGAUCG .......................(((.(.(((((((.((((((((((......-------------.............--......))))...)))))).)))))))...).))) ( -17.91, z-score = -1.06, R) >droYak2.chrX 1405409 116 + 21770863 AUUAACAGCUUCCGUUUUCAAUCCGACCCGUUUCCGAUUCGUUCUCUGGAGAAUAUUCUGCAUUAAAGCAGAAAUUAAAUAGAAAGAAGAGCACAACGAACCGGAAACCACGGUCG .......................(((((.(((((((.((((((((((......((((((((......)))))......)))......))))...)))))).)))))))...))))) ( -31.80, z-score = -3.41, R) >droSec1.super_10 1357870 101 + 3036183 AUUAACCGCUUCCGUUUUCAAUCCGACCCGUUUCCGAUUCGUUCUCUGCAGAA-------------AGCAUAAAAUAAA--ACACGAAGAGCACAACGAACCGGAAACCACGAUCG .......................(((.(.(((((((.((((((((((......-------------.............--......))))...)))))).)))))))...).))) ( -17.91, z-score = -0.96, R) >droSim1.chrX_random 1021371 101 + 5698898 AUUAACCGCUUCCGUUUUCAAUCCGACCCGUUUCCGAUUCGUUCUCUGCAGAA-------------AGCAGAAAAUAAA--ACACGAAGAGCACAACGAACCGGAAACCACGAUCG .......................(((.(.(((((((.((((((.(((((....-------------.))))).......--....(.....)..)))))).)))))))...).))) ( -20.40, z-score = -1.43, R) >consensus AUUAACCGCUUCCGUUUUCAAUCCGACCCGUUUCCGAUUCGUUCUCUGCAGAA_____________AGCAUAAAAUAAA__ACACGAAGAGCACAACGAACCGGAAACCACGAUCG .......................(((.(.(((((((.((((((((((........................................))))...)))))).)))))))...).))) (-17.18 = -17.18 + -0.00)

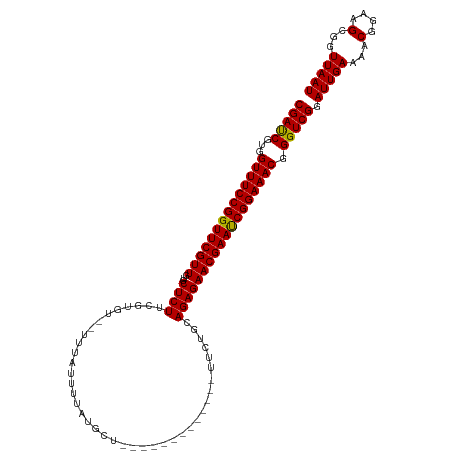
| Location | 1,552,181 – 1,552,282 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.88 |
| Shannon entropy | 0.18222 |
| G+C content | 0.44488 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.38 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.968119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1552181 101 - 22422827 CGAUCGUGGUUUCCGGUUCGUUGUGCUCUUCGUGU--UUUAUUUUAUGCU-------------UUCUGAAGAGAACGAACCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAU (((((...((((((((((((((...(((((((.((--..........)).-------------...))))))))))))))))))))).))))).(((((...((....)).))))) ( -36.50, z-score = -3.12, R) >droEre2.scaffold_4644 1537384 101 - 2521924 CGAUCGUGGUUUCCGGUUCGUUGUGCUCUUAUUUC--UUUAUUAUAUGCU-------------UUCUUCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAU (((((...((((((((((((((...((((......--.............-------------......)))))))))))))))))).))))).(((((...((....)).))))) ( -28.31, z-score = -1.26, R) >droYak2.chrX 1405409 116 - 21770863 CGACCGUGGUUUCCGGUUCGUUGUGCUCUUCUUUCUAUUUAAUUUCUGCUUUAAUGCAGAAUAUUCUCCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCUGUUAAU (((((...((((((((((((((.....................((((((......))))))..(((....))))))))))))))))).))))).(((((...(....)...))))) ( -38.40, z-score = -3.34, R) >droSec1.super_10 1357870 101 - 3036183 CGAUCGUGGUUUCCGGUUCGUUGUGCUCUUCGUGU--UUUAUUUUAUGCU-------------UUCUGCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAU (((((...((((((((((((((.(((.....((((--........)))).-------------....)))...)))))))))))))).))))).(((((...((....)).))))) ( -31.20, z-score = -1.57, R) >droSim1.chrX_random 1021371 101 - 5698898 CGAUCGUGGUUUCCGGUUCGUUGUGCUCUUCGUGU--UUUAUUUUCUGCU-------------UUCUGCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAU (((((...(((((((((((((((..(.....)..)--......((((((.-------------....)))))))))))))))))))).))))).(((((...((....)).))))) ( -35.40, z-score = -2.67, R) >consensus CGAUCGUGGUUUCCGGUUCGUUGUGCUCUUCGUGU__UUUAUUUUAUGCU_____________UUCUGCAGAGAACGAAUCGGAAACGGGUCGGAUUGAAAACGGAAGCGGUUAAU (((((...((((((((((((((...((((........................................)))))))))))))))))).))))).(((((...(....)...))))) (-28.70 = -28.38 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:40 2011