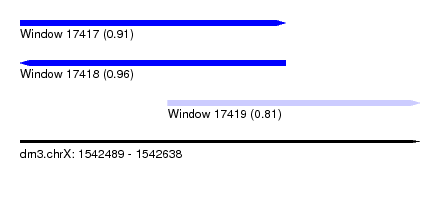
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,542,489 – 1,542,638 |
| Length | 149 |
| Max. P | 0.959067 |

| Location | 1,542,489 – 1,542,588 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 69.48 |
| Shannon entropy | 0.52931 |
| G+C content | 0.66049 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -30.12 |
| Energy contribution | -29.64 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
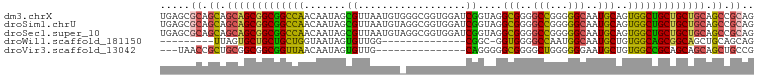
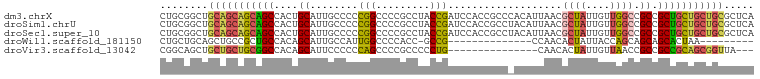
>dm3.chrX 1542489 99 + 22422827 UGAGCGCAGCAGCAGCGGCGGCCAACAAUAGCGUUAAUGUGGGCGGUGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAG .(.(((((((((((((.((.(((.....((.((((......)))).))....)))..(((..(((...)))..))).)).))))))))))).))).... ( -44.40, z-score = -0.48, R) >droSim1.chrU 14847082 99 - 15797150 UGAGCGCAGCAGCAGCGGCGGCCAACAAUAGCGUUAAUGUAGGCGGUGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAG .(.(((((((((((((.((.(((.....((.((((......)))).))....)))..(((..(((...)))..))).)).))))))))))).))).... ( -44.30, z-score = -0.69, R) >droSec1.super_10 1348260 99 + 3036183 UGAGCGCAGCAGCAGCGGCGGCCAACAAUAGCGUUAAUGUAGGCGGUGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAG .(.(((((((((((((.((.(((.....((.((((......)))).))....)))..(((..(((...)))..))).)).))))))))))).))).... ( -44.30, z-score = -0.69, R) >droWil1.scaffold_181150 4589392 75 + 4952429 ---------UUAGUGCUGCUGCUGGUAAUAGUGUUGG--------------CGGC-GGUGGGGCCAAUGGCAAUGCUGUGGCAGCGGCAGCUGCAGCAG ---------...((((((((((((.((.((((((((.--------------((..-((.....))..)).)))))))))).)))))))))).))..... ( -35.70, z-score = -1.73, R) >droVir3.scaffold_13042 5029588 81 + 5191987 ---UAACCGCUGCGGCGGCGGUUAACAAUAGUGUUG---------------CAGGGGGCGGGGCUGGGGGGAAUGCUGUGGCCGCAGCAGCAGCUGCCG ---.....((((((((.(((((...(..((((.(((---------------(.....)))).))))..).....))))).)))))))).((....)).. ( -37.30, z-score = -1.27, R) >consensus UGAGCGCAGCAGCAGCGGCGGCCAACAAUAGCGUUAAUGU_GGCGGUGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAG .....((.((.(((((((((((((((......)))......................(((..(((...)))..)))...)))))))))))).)).)).. (-30.12 = -29.64 + -0.48)
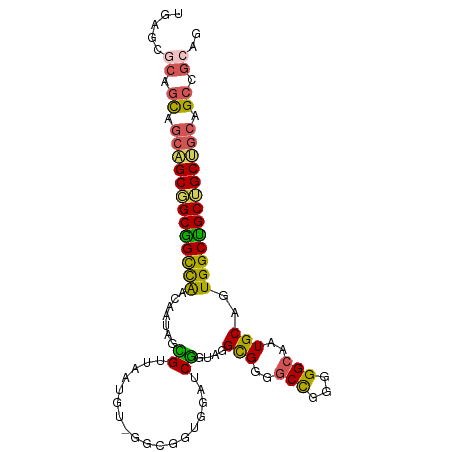
| Location | 1,542,489 – 1,542,588 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 69.48 |
| Shannon entropy | 0.52931 |
| G+C content | 0.66049 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.64 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1542489 99 - 22422827 CUGCGGCUGCAGCAGCAGCCACUGCAUUGCCCCCGGCCCCGCCUACCGAUCCACCGCCCACAUUAACGCUAUUGUUGGCCGCCGCUGCUGCUGCGCUCA ....(((.(((((((((((....((.........(((...)))............(((.(((.((....)).))).))).)).)))))))))))))).. ( -36.20, z-score = -1.30, R) >droSim1.chrU 14847082 99 + 15797150 CUGCGGCUGCAGCAGCAGCCACUGCAUUGCCCCCGGCCCCGCCUACCGAUCCACCGCCUACAUUAACGCUAUUGUUGGCCGCCGCUGCUGCUGCGCUCA ....(((.(((((((((((....((.........(((...)))............(((.(((.((....)).))).))).)).)))))))))))))).. ( -36.20, z-score = -1.34, R) >droSec1.super_10 1348260 99 - 3036183 CUGCGGCUGCAGCAGCAGCCACUGCAUUGCCCCCGGCCCCGCCUACCGAUCCACCGCCUACAUUAACGCUAUUGUUGGCCGCCGCUGCUGCUGCGCUCA ....(((.(((((((((((....((.........(((...)))............(((.(((.((....)).))).))).)).)))))))))))))).. ( -36.20, z-score = -1.34, R) >droWil1.scaffold_181150 4589392 75 - 4952429 CUGCUGCAGCUGCCGCUGCCACAGCAUUGCCAUUGGCCCCACC-GCCG--------------CCAACACUAUUACCAGCAGCAGCACUAA--------- .((((((.((((..((((...)))).......(((((......-...)--------------)))).........)))).))))))....--------- ( -24.20, z-score = -1.61, R) >droVir3.scaffold_13042 5029588 81 - 5191987 CGGCAGCUGCUGCUGCGGCCACAGCAUUCCCCCCAGCCCCGCCCCCUG---------------CAACACUAUUGUUAACCGCCGCCGCAGCGGUUA--- ....(((((((((.(((((.....................((.....)---------------)((((....))))....))))).))))))))).--- ( -27.60, z-score = -1.21, R) >consensus CUGCGGCUGCAGCAGCAGCCACUGCAUUGCCCCCGGCCCCGCCUACCGAUCCACCGCC_ACAUUAACGCUAUUGUUGGCCGCCGCUGCUGCUGCGCUCA ........(((((((((((....((........(((.........)))...................((((....)))).)).)))))))))))..... (-21.48 = -21.64 + 0.16)
| Location | 1,542,544 – 1,542,638 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Shannon entropy | 0.33678 |
| G+C content | 0.71683 |
| Mean single sequence MFE | -53.34 |
| Consensus MFE | -40.26 |
| Energy contribution | -41.14 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
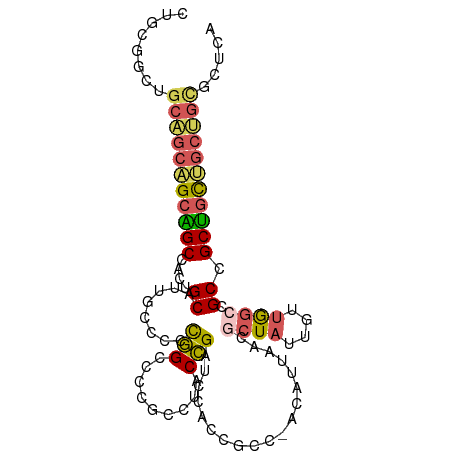
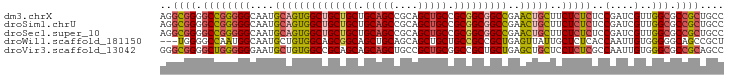
>dm3.chrX 1542544 94 + 22422827 AGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGCUUCUCUCUCCGAUCGUUGGCGCCGCUGCC .(((((.((((((((....((((((((((((((.(((((....))))).)))))))))..)))))....)))))(....)..))).)))))... ( -54.20, z-score = -1.67, R) >droSim1.chrU 14847137 94 - 15797150 AGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGCUUCUCUCUCCGAUCGUUGGCGCCGCUGCC .(((((.((((((((....((((((((((((((.(((((....))))).)))))))))..)))))....)))))(....)..))).)))))... ( -54.20, z-score = -1.67, R) >droSec1.super_10 1348315 94 + 3036183 AGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGCUUCUCUCUCCGAUCGUUGGCGCCGCUGCC .(((((.((((((((....((((((((((((((.(((((....))))).)))))))))..)))))....)))))(....)..))).)))))... ( -54.20, z-score = -1.67, R) >droWil1.scaffold_181150 4589426 91 + 4952429 ---UGGGGCCAAUGGCAAUGCUGUGGCAGCGGCAGCUGCAGCAGCUGCUGCCGCCGCUGAGUUAUUGCUCUCACCAAUUGUGGGGGCAGCCGCU ---.(.(((.((((((...((.(((((((((((.((....)).))))))))))).))...))))))((((((((.....)))))))).))).). ( -49.50, z-score = -1.89, R) >droVir3.scaffold_13042 5029625 94 + 5191987 GGGCGGGGCUGGGGGGAAUGCUGUGGCCGCAGCAGCAGCUGCCGCUGCGGCCGCUGCUGAGCUGCUCCUCUCGCCAAUUGUGGGCGCCGCAGCC .((((.(((.(((((((..((((((((((((((.((....)).))))))))))).....)))...)))))))(((.......)))))).).))) ( -54.60, z-score = -0.81, R) >consensus AGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGCUUCUCUCUCCGAUCGUUGGCGCCGCUGCC ..((((.((((((((....((((((((((((((.(((((....))))).)))))))))..)))))..)))))..(....)..))).)))).... (-40.26 = -41.14 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:36 2011