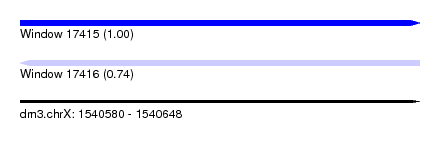
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,540,580 – 1,540,648 |
| Length | 68 |
| Max. P | 0.999610 |

| Location | 1,540,580 – 1,540,648 |
|---|---|
| Length | 68 |
| Sequences | 7 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 61.29 |
| Shannon entropy | 0.81185 |
| G+C content | 0.48090 |
| Mean single sequence MFE | -17.63 |
| Consensus MFE | -12.53 |
| Energy contribution | -13.84 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
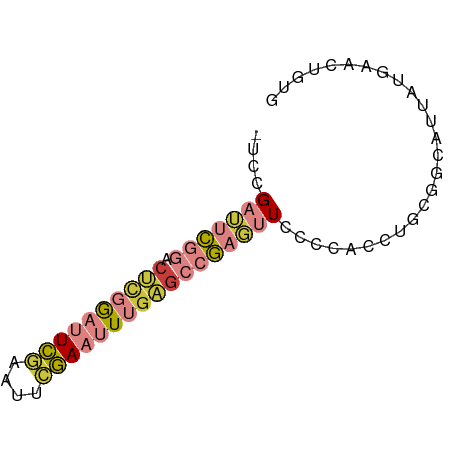
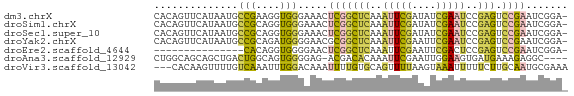
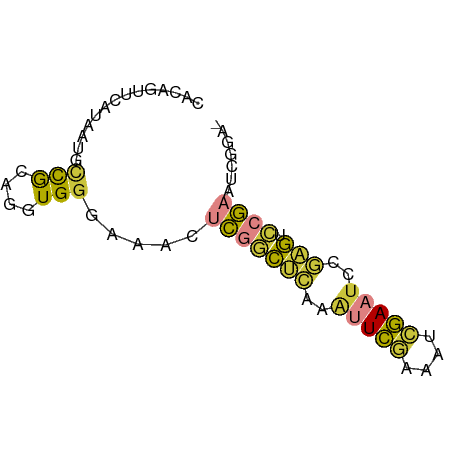
>dm3.chrX 1540580 68 + 22422827 -UCCGAUUCGGACUCGGAUUCGAUAUCGAAUUUGAGCCGAGUUUCCCACCUUCGGCAUUAUGAACUGUG -...(((((((.((((((((((....)))))))))))))))))...(((.((((......))))..))) ( -23.70, z-score = -3.09, R) >droSim1.chrX 1119485 68 + 17042790 -UCCGAUUCGGACUCGGAUUCGAUAUCGAAUUUGAGCCGAGUUUCCCACCUGCGGCAUUAUGAACUGUG -...(((((((.((((((((((....)))))))))))))))))........((((.........)))). ( -23.00, z-score = -2.69, R) >droSec1.super_10 1346356 68 + 3036183 -UCCGAUUCGGACUCGGAUUCGAUAUCGAAUUUGAGCCGAGUUUCCCACCUGCGGCAUUAUGAACUGUG -...(((((((.((((((((((....)))))))))))))))))........((((.........)))). ( -23.00, z-score = -2.69, R) >droYak2.chrX 1393266 68 + 21770863 -UCCGAUUCGGACUCGGAUUCGAAUUCGAAUUUGAGCCGCGUUCCCCAUCUGCGGCAUUAUGAACUGUG -((((...)))).(((((((((....)))))))))((((((.........))))))............. ( -21.40, z-score = -1.69, R) >droEre2.scaffold_4644 1525179 53 + 2521924 -UCCGAUUCGGACUCGGAGUCGAAUUCGAAUUUGAGCCGAGUUCCCCACCUGUG--------------- -...(((((((.((((((.(((....))).)))))))))))))...........--------------- ( -18.20, z-score = -2.22, R) >droAna3.scaffold_12929 1410498 64 + 3277472 ----GCCUCUUUCAUCACUUCCAAUUCGAAUUUGUGUCGU-CUCCCCACUGCCAGUCAGCUGCUGCCAG ----...........((((((......)))...)))....-.......(((.((((.....)))).))) ( -5.50, z-score = 0.77, R) >droVir3.scaffold_13042 5027537 66 + 5191987 UUUCGCAUUGCAAGAAAAAUUUACUUAAAACUGCACAAAAUUUGUCCAAAUUUGACAAAACUUGUG--- ((((.........))))................(((((..((((((.......))))))..)))))--- ( -8.60, z-score = -0.67, R) >consensus _UCCGAUUCGGACUCGGAUUCGAAUUCGAAUUUGAGCCGAGUUCCCCACCUGCGGCAUUAUGAACUGUG ....(((((((.((((((((((....))))))))))))))))).......................... (-12.53 = -13.84 + 1.31)

| Location | 1,540,580 – 1,540,648 |
|---|---|
| Length | 68 |
| Sequences | 7 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 61.29 |
| Shannon entropy | 0.81185 |
| G+C content | 0.48090 |
| Mean single sequence MFE | -15.83 |
| Consensus MFE | -7.37 |
| Energy contribution | -7.01 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.73 |
| Mean z-score | -0.40 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1540580 68 - 22422827 CACAGUUCAUAAUGCCGAAGGUGGGAAACUCGGCUCAAAUUCGAUAUCGAAUCCGAGUCCGAAUCGGA- ..............((((.....(....)(((((((..(((((....)))))..))).)))).)))).- ( -20.40, z-score = -1.54, R) >droSim1.chrX 1119485 68 - 17042790 CACAGUUCAUAAUGCCGCAGGUGGGAAACUCGGCUCAAAUUCGAUAUCGAAUCCGAGUCCGAAUCGGA- ..............(((......(....)(((((((..(((((....)))))..))).))))..))).- ( -17.90, z-score = -0.62, R) >droSec1.super_10 1346356 68 - 3036183 CACAGUUCAUAAUGCCGCAGGUGGGAAACUCGGCUCAAAUUCGAUAUCGAAUCCGAGUCCGAAUCGGA- ..............(((......(....)(((((((..(((((....)))))..))).))))..))).- ( -17.90, z-score = -0.62, R) >droYak2.chrX 1393266 68 - 21770863 CACAGUUCAUAAUGCCGCAGAUGGGGAACGCGGCUCAAAUUCGAAUUCGAAUCCGAGUCCGAAUCGGA- .............(((((...........)))))((..(((((.(((((....))))).)))))..))- ( -18.40, z-score = -0.37, R) >droEre2.scaffold_4644 1525179 53 - 2521924 ---------------CACAGGUGGGGAACUCGGCUCAAAUUCGAAUUCGACUCCGAGUCCGAAUCGGA- ---------------....(((..(((.(((((.......(((....)))..))))))))..)))...- ( -14.10, z-score = 0.10, R) >droAna3.scaffold_12929 1410498 64 - 3277472 CUGGCAGCAGCUGACUGGCAGUGGGGAG-ACGACACAAAUUCGAAUUGGAAGUGAUGAAAGAGGC---- ((..((.(((((....))).(((.(...-.)..)))................)).))..))....---- ( -10.20, z-score = 0.75, R) >droVir3.scaffold_13042 5027537 66 - 5191987 ---CACAAGUUUUGUCAAAUUUGGACAAAUUUUGUGCAGUUUUAAGUAAAUUUUUCUUGCAAUGCGAAA ---.(((((.((((((.......)))))).)))))(((....((((.........))))...))).... ( -11.90, z-score = -0.49, R) >consensus CACAGUUCAUAAUGCCGCAGGUGGGAAACUCGGCUCAAAUUCGAAAUCGAAUCCGAGUCCGAAUCGGA_ ..............(((....))).....(((((((..(((((....)))))..))).))))....... ( -7.37 = -7.01 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:33 2011