
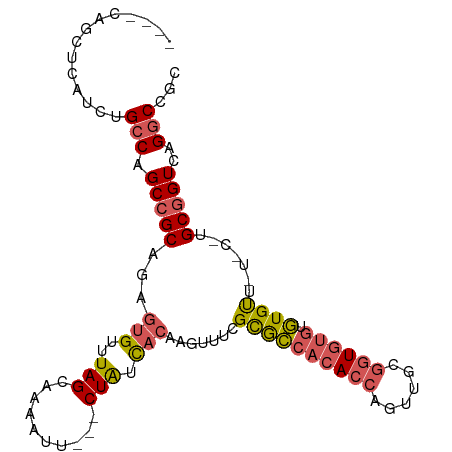
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,532,602 – 1,532,713 |
| Length | 111 |
| Max. P | 0.728649 |

| Location | 1,532,602 – 1,532,704 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.27 |
| Shannon entropy | 0.39035 |
| G+C content | 0.53484 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -20.68 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1532602 102 + 22422827 GCUUCAGCUCAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU---CUAUCACAAGUUUCGCGCCACACCAGUUGCGGUGUGUGUGUUCUGCAUGCGGUCAGGCCGC ((....))......(((.((((((..(((..(((.......---))).)))..((...((((((((((......)))))).))))...)).))))))..)))... ( -32.80, z-score = -0.76, R) >droSec1.super_10 1338342 102 + 3036183 GCUCCAGCUCAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU---CUAUCACAAGUUUCGCACCACACCAGUUGCGGUGUGUGUGUUCUGCGUGCGGUCAGGCCGC ((....))...((((((.(((((((((((..(((.......---))).))).......((((((((((......)))))).)))))))))).)))).)))).... ( -35.80, z-score = -1.86, R) >droYak2.chrX 1385264 92 + 21770863 ----CAGCUCAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU---CUAUCACAAGUUUCGCACCACACCAGUUGCGGUGUGUGUGCU------GCGGUCAGGCCGC ----..........(((.((((((..(((..(((.......---))).))).......((((((((((......)))))).)))))------)))))..)))... ( -33.00, z-score = -1.96, R) >droEre2.scaffold_4644 1517223 92 + 2521924 ----CAGCUCAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU---CUAUCACAAGUUUCGCGCCACACCAGUUGCGGUGUGUGUGCU------GCGGUCAGGCCGC ----..........(((.((((((..(((..(((.......---))).))).......((((((((((......)))))).)))))------)))))..)))... ( -32.60, z-score = -1.49, R) >droGri2.scaffold_15081 3680413 94 + 4274704 ----CAGCUUG---GGCAGCAGCAG--UGUUUAGCAAAAAUGCACUGUUUUUGAAUAUGUGGCAAAC-AUUUUAAUUGCCGCUGUUAUCC-UGCGGUUAGGCCAC ----..(((((---(.(.(((((((--(((...........))))))......((((.(((((((..-.......)))))))))))...)-)))).))))))... ( -27.60, z-score = -0.39, R) >consensus ____CAGCUCAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU___CUAUCACAAGUUUCGCGCCACACCAGUUGCGGUGUGUGUGUU_U_C_UGCGGUCAGGCCGC ..............(((.(((((...(((..(((..........))).))).......((((((((((......)))))).)))).......)))))..)))... (-20.68 = -21.20 + 0.52)


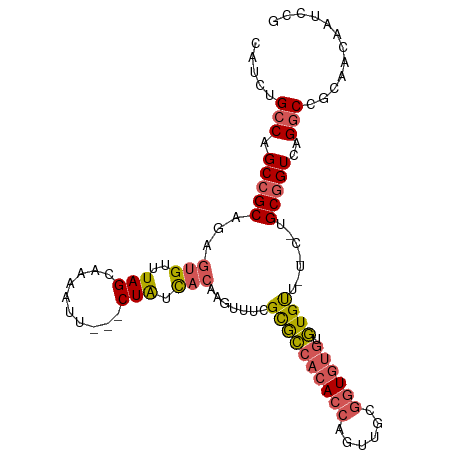
| Location | 1,532,611 – 1,532,713 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.37255 |
| G+C content | 0.51506 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -20.68 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1532611 102 + 22422827 CAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU---CUAUCACAAGUUUCGCGCCACACCAGUUGCGGUGUGUGUGUUCUGCAUGCGGUCAGGCCGCAACAAUCCG ............((((((((..(((.......---))).))).......((((((((((......)))))).))))))))).(((((.....)))))........ ( -32.30, z-score = -1.26, R) >droSec1.super_10 1338351 102 + 3036183 CAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU---CUAUCACAAGUUUCGCACCACACCAGUUGCGGUGUGUGUGUUCUGCGUGCGGUCAGGCCGCAACAAUCCG ..((((((.(((((((((((..(((.......---))).))).......((((((((((......)))))).)))))))))).)))).))))............. ( -35.10, z-score = -2.27, R) >droYak2.chrX 1385269 95 + 21770863 CAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU---CUAUCACAAGUUUCGCACCACACCAGUUGCGGUGUGUGUGCU------GCGGUCAGGCCGCAACAAUCC- .....(((.((((((..(((..(((.......---))).))).......((((((((((......)))))).)))))------)))))..)))...........- ( -33.00, z-score = -2.44, R) >droEre2.scaffold_4644 1517228 96 + 2521924 CAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU---CUAUCACAAGUUUCGCGCCACACCAGUUGCGGUGUGUGUGCU------GCGGUCAGGCCGCAACAAUUCG .....(((.((((((..(((..(((.......---))).))).......((((((((((......)))))).)))))------)))))..)))............ ( -32.60, z-score = -1.73, R) >droGri2.scaffold_15081 3680418 98 + 4274704 ---UGGGCAGCAGCAG--UGUUUAGCAAAAAUGCACUGUUUUUGAAUAUGUGGCAAAC-AUUUUAAUUGCCGCUGUUAU-CCUGCGGUUAGGCCACAACAAUUUG ---((..(((.(((((--(((...........)))))))).)))..))((((((....-.........(((((......-...)))))...))))))........ ( -26.89, z-score = -0.27, R) >consensus CAUCUGCCAGCCGCAGAGUGUUUAGCAAAAUU___CUAUCACAAGUUUCGCGCCACACCAGUUGCGGUGUGUGUGUU_U_C_UGCGGUCAGGCCGCAACAAUCCG .....(((.(((((...(((..(((..........))).))).......((((((((((......)))))).)))).......)))))..)))............ (-20.68 = -21.20 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:30 2011