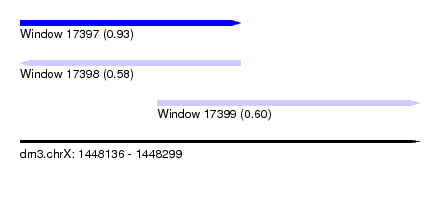
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,448,136 – 1,448,299 |
| Length | 163 |
| Max. P | 0.934216 |

| Location | 1,448,136 – 1,448,226 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.51662 |
| G+C content | 0.54546 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -17.77 |
| Energy contribution | -16.82 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.934216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
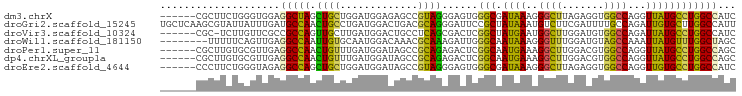
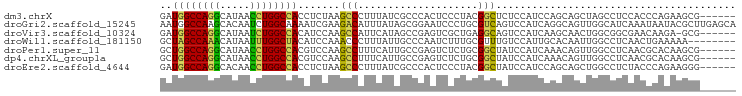
>dm3.chrX 1448136 90 + 22422827 ------CGCUUCUGGGUGGAGGCUAGCUGCUGGAUGGAGAGCCGUAGGGAGUGGGCGAUAAAGGGCUUAGAGGUGGCCAGGUUAUGCCUGGCCAUC ------.((((((((((....(((.(((.((..((((....)))).)).))).)))........))))))))))(((((((.....)))))))... ( -38.10, z-score = -2.38, R) >droGri2.scaffold_15245 15619433 96 + 18325388 UGCUCAAGCGUAUUAUUUGAUGCCAACUGCCUGAUGGACUGACGCAGGGAUUCCGCUAUAAAUGUCUUCGAUUUUGCCAGAUUGUGCUUGGCCAUU .((.((((((((((....)))))(((((((..(((.((..(((((.((....)))).......))).)).)))..).))).))).))))))).... ( -20.71, z-score = 0.84, R) >droVir3.scaffold_10324 1287919 89 - 1288806 ------CGC-UCUUGUUCGCCGCCAGUUGCUUGAUGGACUGCCUCAGCGACUCGGCUAUGAAUGGCUUGGAUGUGGCCAGAUUAUGCCUGGCCAUC ------.((-(...(((((..((((((((((.((.((....))))))))))).)))..))))))))......((((((((.......)))))))). ( -35.70, z-score = -2.39, R) >droWil1.scaffold_181150 4438725 88 + 4952429 --------UUUUUCAGUUGAGGCCAAUUGUGCAAUGGACAAACGCAAAGAUUGGGCAAUAAAGGGUUUGGAUGUAGCCAAAUUAUGUUUGGCUAGC --------.......(((.(((((.(((((.((((..............)))).)))))....))))).))).(((((((((...))))))))).. ( -22.84, z-score = -1.23, R) >droPer1.super_11 1710578 90 - 2846995 ------CGCUUGUGCGUUGAGGCCAACUGUUUGAUGGAUAGCCGCAGAGACUCGGCAAUGAAAGGCUUGGACGUGGCCAGGUUAUGCCUGGCCAGC ------.(((...(((((.(((((...((((.....))))((((.((...)))))).......))))).)))))(((((((.....)))))))))) ( -37.90, z-score = -2.56, R) >dp4.chrXL_group1a 4600546 90 - 9151740 ------CGCUUGUGCGUUGAGGCCAACUGUUUGAUGGAUAGCCGCAGAGACUCGGCAAUGAAAGGCUUGGACGUGGCCAGGUUAUGCCUGGCCAGC ------.(((...(((((.(((((...((((.....))))((((.((...)))))).......))))).)))))(((((((.....)))))))))) ( -37.90, z-score = -2.56, R) >droEre2.scaffold_4644 1440558 90 + 2521924 ------CCCUUCUGGGUAGAGGCCAGCUGCUGGAUGGAUAGCCGUAGGGAGUGGGCGAUAAAGGGCUUAGAGGUGGCCAGGUUGUGCCUGGCCAUC ------..(((((((((....(((.(((.((..((((....)))).)).))).)))........)))))))))((((((((.....)))))))).. ( -39.60, z-score = -1.91, R) >consensus ______CGCUUCUGGGUUGAGGCCAACUGCUUGAUGGACAGCCGCAGAGACUCGGCAAUAAAGGGCUUGGAUGUGGCCAGGUUAUGCCUGGCCAUC ....................(((((.((((.............))))......(((.((((..((((.......))))...))))))))))))... (-17.77 = -16.82 + -0.95)
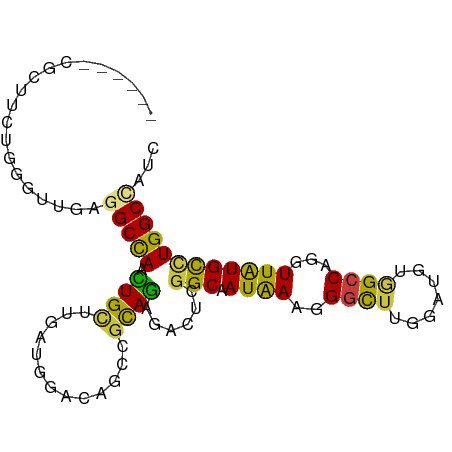
| Location | 1,448,136 – 1,448,226 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.51662 |
| G+C content | 0.54546 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -13.86 |
| Energy contribution | -13.06 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1448136 90 - 22422827 GAUGGCCAGGCAUAACCUGGCCACCUCUAAGCCCUUUAUCGCCCACUCCCUACGGCUCUCCAUCCAGCAGCUAGCCUCCACCCAGAAGCG------ ..((((((((.....))))))))..............................((((((......)).)))).((.((......)).)).------ ( -21.10, z-score = -0.86, R) >droGri2.scaffold_15245 15619433 96 - 18325388 AAUGGCCAAGCACAAUCUGGCAAAAUCGAAGACAUUUAUAGCGGAAUCCCUGCGUCAGUCCAUCAGGCAGUUGGCAUCAAAUAAUACGCUUGAGCA ....(((((((.....(((((......(((....)))...((((.....)))))))))........((.....))............))))).)). ( -20.60, z-score = 0.37, R) >droVir3.scaffold_10324 1287919 89 + 1288806 GAUGGCCAGGCAUAAUCUGGCCACAUCCAAGCCAUUCAUAGCCGAGUCGCUGAGGCAGUCCAUCAAGCAACUGGCGGCGAACAAGA-GCG------ ..((((((((.....)))))))).......((..((....(((((((.(((((((....)).)).))).)))..))))....))..-)).------ ( -27.90, z-score = -0.88, R) >droWil1.scaffold_181150 4438725 88 - 4952429 GCUAGCCAAACAUAAUUUGGCUACAUCCAAACCCUUUAUUGCCCAAUCUUUGCGUUUGUCCAUUGCACAAUUGGCCUCAACUGAAAAA-------- ..((((((((.....))))))))...........(((((((.(((((...((((.........))))..)))))...))).))))...-------- ( -15.10, z-score = -0.93, R) >droPer1.super_11 1710578 90 + 2846995 GCUGGCCAGGCAUAACCUGGCCACGUCCAAGCCUUUCAUUGCCGAGUCUCUGCGGCUAUCCAUCAAACAGUUGGCCUCAACGCACAAGCG------ ((((((((((.....)))))))........((........((((........)))).............((((....))))))...))).------ ( -28.30, z-score = -1.56, R) >dp4.chrXL_group1a 4600546 90 + 9151740 GCUGGCCAGGCAUAACCUGGCCACGUCCAAGCCUUUCAUUGCCGAGUCUCUGCGGCUAUCCAUCAAACAGUUGGCCUCAACGCACAAGCG------ ((((((((((.....)))))))........((........((((........)))).............((((....))))))...))).------ ( -28.30, z-score = -1.56, R) >droEre2.scaffold_4644 1440558 90 - 2521924 GAUGGCCAGGCACAACCUGGCCACCUCUAAGCCCUUUAUCGCCCACUCCCUACGGCUAUCCAUCCAGCAGCUGGCCUCUACCCAGAAGGG------ ..((((((((.....))))))))........((((((...(((..........))).......((((...))))..........))))))------ ( -27.30, z-score = -1.17, R) >consensus GAUGGCCAGGCAUAACCUGGCCACAUCCAAGCCCUUUAUUGCCGAGUCCCUGCGGCUAUCCAUCAAGCAGUUGGCCUCAACCCAGAAGCG______ ..((((((((.....)))))))).......(((....................)))........................................ (-13.86 = -13.06 + -0.79)
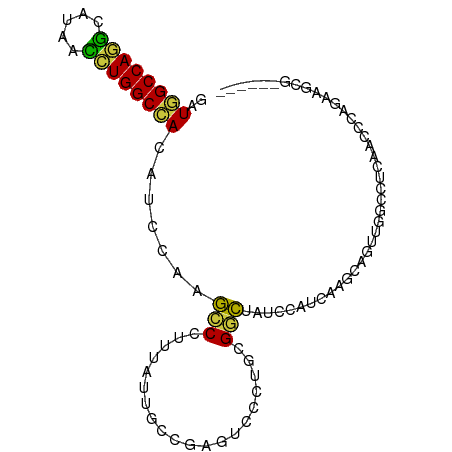
| Location | 1,448,192 – 1,448,299 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 60.30 |
| Shannon entropy | 0.54113 |
| G+C content | 0.58837 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -20.39 |
| Energy contribution | -21.60 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
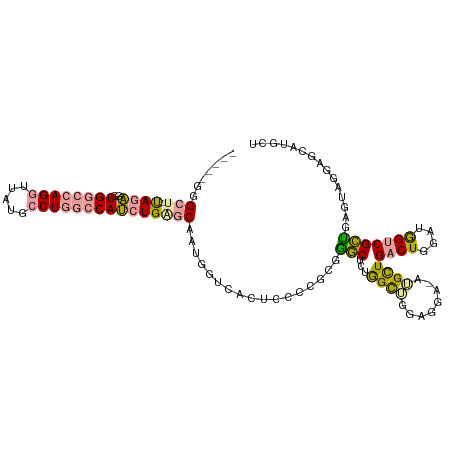
>dm3.chrX 1448192 107 + 22422827 -----GGGCU-UAGAGGUGGCCAGGUUAUGCCUGGCCAUCUGGGCAAUGGUCACUCCCCGCGAGCGCUGGCUGGAAGA-AGGCUGACUGGAUGGUCGCUGAGUAGGAGCAUGCU -----..(((-((((..((((((((.....))))))))))))))).(((.((((((...((((.(((..(..((....-...))..)..).)).)))).))))..)).)))... ( -44.10, z-score = -0.31, R) >droAna3.scaffold_13266 15970207 98 - 19884421 UGUGGGGGCUACAGUGGUGGU-AGAUGUUGCCU---CAGAUGU-CGAUGAACUCGAUGUGGGGACUGCAGUGGACGUAGAUGUUGCCUCUGUAGUCGUCGAAC----------- ..(((.((((((((.(((..(-(.(((((.(((---((..(((-(((.....)))))))))))((....)).)))))...))..))).)))))))).)))...----------- ( -35.30, z-score = -1.91, R) >droEre2.scaffold_4644 1440614 107 + 2521924 -----GGGCU-UAGAGGUGGCCAGGUUGUGCCUGGCCAUCUGAGCAAUGGUCACUCCUCGCGGGCUCUGGCUGGAGGA-AUGCUGACUGGAUGGUCGCUGAGUAGGAGCAUGCU -----..(((-((((..((((((((.....))))))))))))))).(((.((.((.(((((((.(((..(..((....-...))..)..)).).)))).))).)))).)))... ( -47.50, z-score = -1.52, R) >consensus _____GGGCU_UAGAGGUGGCCAGGUUAUGCCUGGCCAUCUGAGCAAUGGUCACUCCCCGCGGGCUCUGGCUGGAGGA_AUGCUGACUGGAUGGUCGCUGAGUAGGAGCAUGCU ......(((.....(((((((((((.....))))))))))).......(((...(((((...(((....))).)))))...)))((((....)))))))............... (-20.39 = -21.60 + 1.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:20 2011