
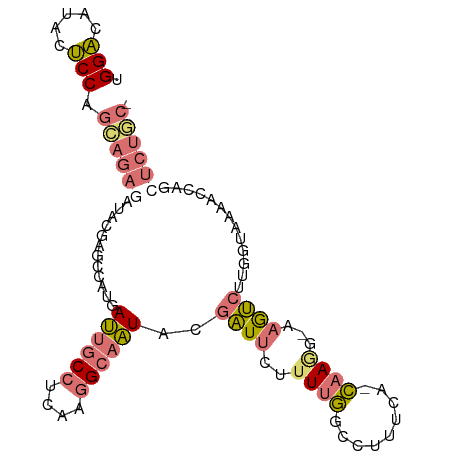
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,412,604 – 1,412,701 |
| Length | 97 |
| Max. P | 0.637089 |

| Location | 1,412,604 – 1,412,701 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 63.28 |
| Shannon entropy | 0.60223 |
| G+C content | 0.47857 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -10.59 |
| Energy contribution | -12.15 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.637089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1412604 97 + 22422827 AGGUGUUGAGCCAGGAGAAUUACGCUCC-CAACUCCGCGCCAGCUGUGCAGUUGUUUCCCCUAACCUGGAAGGUAUGCCAACUCAAUCUACGGUCUCC- .(((((.(((...((((.......))))-...))).))))).((((((..((((.((((........))))((....))....)))).))))))....- ( -26.20, z-score = -0.29, R) >droYak2.chrU 8888291 89 - 28119190 UGGA-ACACUCCAGCAGAGAUACGAGCUAUGAUUGCCUCAAGGCAAUACGAUUCUCUUGGCCUUUCGGCAAUGCAAAUCUUGGUAAAACC--------- ((((-....))))((.(((((..(((...((((((((....)))))).))...)))...(((....))).......))))).))......--------- ( -22.10, z-score = -1.05, R) >droSec1.super_1989 535 97 - 3922 UGGACAUACUCCAGCAGAGAUACGAGCCAUGAUUGCCUCAAGGCAAUACGAUUCUUUUGGCCUUUCA-CAAGG-AAGUCUUGGUAAAACCAGCUCUGCU ((((.....))))((((((......((((..((((((....))))))..((((..((((........-)))).-.)))).))))........)))))). ( -29.24, z-score = -2.12, R) >droSim1.chr2L 20995059 97 + 22036055 UGGACAUACUCCAGCAGAGAUACGAGCUAUGAUUGCCUCAAGGCAAUAAGAUUCUUUUGGCCUUUCA-CAAAG-AAGUCUUGGUAACACCAGCUCUGCU ((((.....))))((((((............((((((....))))))((((((..((((........-)))).-.))))))((.....))..)))))). ( -28.90, z-score = -2.18, R) >consensus UGGACAUACUCCAGCAGAGAUACGAGCCAUGAUUGCCUCAAGGCAAUACGAUUCUUUUGGCCUUUCA_CAAGG_AAGUCUUGGUAAAACCAGCUCUGC_ .(((.....))).((((((............((((((....))))))..((((......((......))......)))).............)))))). (-10.59 = -12.15 + 1.56)


| Location | 1,412,604 – 1,412,701 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 63.28 |
| Shannon entropy | 0.60223 |
| G+C content | 0.47857 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -11.30 |
| Energy contribution | -12.11 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.531272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1412604 97 - 22422827 -GGAGACCGUAGAUUGAGUUGGCAUACCUUCCAGGUUAGGGGAAACAACUGCACAGCUGGCGCGGAGUUG-GGAGCGUAAUUCUCCUGGCUCAACACCU -(....).........((((((((..(((........)))(....)...))).)))))((.(..((((..-((((.......))))..))))..).)). ( -31.50, z-score = -0.49, R) >droYak2.chrU 8888291 89 + 28119190 ---------GGUUUUACCAAGAUUUGCAUUGCCGAAAGGCCAAGAGAAUCGUAUUGCCUUGAGGCAAUCAUAGCUCGUAUCUCUGCUGGAGUGU-UCCA ---------((..(..((((((..(((...(((....)))...(((....(.((((((....)))))))....))))))..)))..)))...).-.)). ( -26.10, z-score = -1.43, R) >droSec1.super_1989 535 97 + 3922 AGCAGAGCUGGUUUUACCAAGACUU-CCUUG-UGAAAGGCCAAAAGAAUCGUAUUGCCUUGAGGCAAUCAUGGCUCGUAUCUCUGCUGGAGUAUGUCCA (((((((.(((((((.(((((....-.))))-.).)))))))...((..(((((((((....)))))).)))..))....)))))))(((.....))). ( -34.30, z-score = -2.85, R) >droSim1.chr2L 20995059 97 - 22036055 AGCAGAGCUGGUGUUACCAAGACUU-CUUUG-UGAAAGGCCAAAAGAAUCUUAUUGCCUUGAGGCAAUCAUAGCUCGUAUCUCUGCUGGAGUAUGUCCA (((((((.(((.((((..((((.((-((((.-((......))))))))))))((((((....))))))..)))).).)).)))))))(((.....))). ( -27.40, z-score = -0.88, R) >consensus _GCAGAGCUGGUUUUACCAAGACUU_CCUUG_UGAAAGGCCAAAAGAAUCGUAUUGCCUUGAGGCAAUCAUAGCUCGUAUCUCUGCUGGAGUAUGUCCA .((((((..((.....))..........(((((..(((((...............)))))..))))).............)))))).(((.....))). (-11.30 = -12.11 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:15 2011