
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,626,885 – 9,626,981 |
| Length | 96 |
| Max. P | 0.893257 |

| Location | 9,626,885 – 9,626,981 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 67.12 |
| Shannon entropy | 0.61991 |
| G+C content | 0.49244 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -9.99 |
| Energy contribution | -14.05 |
| Covariance contribution | 4.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.694850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
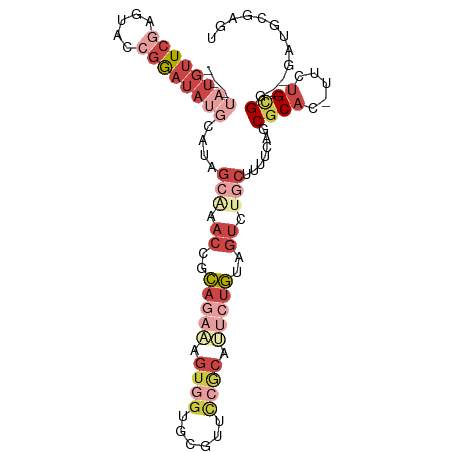
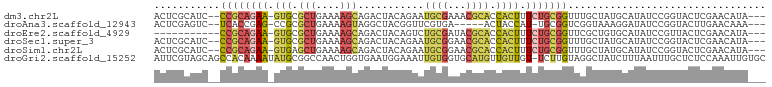
>dm3.chr2L 9626885 96 + 23011544 ---UAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGUGGUGCGUUUCGCAUUCUGUAGUCUGCUUUUCAGCGCAC-UUCUGCGG--GAUGCGAGU ---((((((((.....))))))))....((..(((((.....)))))))..((((((((((((((..(((........))).-..))))))--)))))))). ( -36.10, z-score = -2.50, R) >droAna3.scaffold_12943 1051887 90 + 5039921 ---UUUGUUCAAGUACCGGAUAUCCUUUACCGACCGCA-AUGGUAGU-----UCACGAACCGUAGCCUACUUUUCAGCGCGG-CUCGGUGA--GACUCGAGU ---....(((.(((...((....))((((((((((((.-(((((.(.-----...)..))))).((..........))))))-.)))))))--)))).))). ( -25.40, z-score = -0.89, R) >droEre2.scaffold_4929 10233704 87 + 26641161 ---UAUGUUCGAGUAACGGAUAUGCACAGCGAACCGCAGAAAGUGGUGCGUAUCGCAGACUGUAGUCUGCUUUUCAGCGCAC-UUCUGCGG----------- ---((((((((.....)))))))).........(((((((....(((((((...((((((....))))))......))))))-))))))))----------- ( -35.20, z-score = -3.08, R) >droSec1.super_3 5073346 96 + 7220098 ---UAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGUGGUGCGUUCCGCAUUCUGUAGUCUGCUUUUCAGCGCAC-UUCUGCGG--GAUGCGAGU ---((((((((.....))))))))....(((..(((((((....((((((((..(((..(....)..))).....)))))))-))))))))--..))).... ( -35.70, z-score = -2.29, R) >droSim1.chr2L 9399619 96 + 22036055 ---UAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGUGGUGCGUUCCGCAUUCUGUAGUCUGCUUUUCAGCUCAC-UUCUGCGG--GAUGCGAGU ---((((((((.....))))))))....(((..((((((((.((((.((.....(((..(....)..)))......))))))-))))))))--..))).... ( -34.10, z-score = -2.19, R) >droGri2.scaffold_15252 13654200 101 - 17193109 GCACAAUUUGGAGAGCAAAUUAAAGAUAGCCUACAAGA-ACAACAACAUGCACCACAAUUUCCAUUCACCAGUUGGCCGCAUAUUUUGUGGCUGCUACGAAU .....(((((...((((.....................-....((((........................))))((((((.....)))))))))).))))) ( -17.16, z-score = 0.18, R) >consensus ___UAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGUGGUGCGUUCCGCAUUCUGUAGUCUGCUUUUCAGCGCAC_UUCUGCGG__GAUGCGAGU ...((((((((.....))))))))....(((.((..(((((.((((......)))).)))))..)).))).......((((.....))))............ ( -9.99 = -14.05 + 4.06)

| Location | 9,626,885 – 9,626,981 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 67.12 |
| Shannon entropy | 0.61991 |
| G+C content | 0.49244 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -11.00 |
| Energy contribution | -12.37 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
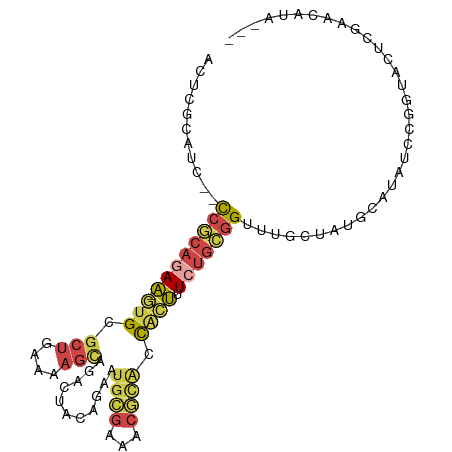
>dm3.chr2L 9626885 96 - 23011544 ACUCGCAUC--CCGCAGAA-GUGCGCUGAAAAGCAGACUACAGAAUGCGAAACGCACCACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUA--- ....(((..--((((((((-(((.(((....)))...........((((...)))).))).))))))))..))).........................--- ( -29.80, z-score = -2.40, R) >droAna3.scaffold_12943 1051887 90 - 5039921 ACUCGAGUC--UCACCGAG-CCGCGCUGAAAAGUAGGCUACGGUUCGUGA-----ACUACCAU-UGCGGUCGGUAAAGGAUAUCCGGUACUUGAACAAA--- ..((((((.--.((((((.-(((((.((....((((..((((...)))).-----.)))))).-))))))))))...((....)))..)))))).....--- ( -30.80, z-score = -2.10, R) >droEre2.scaffold_4929 10233704 87 - 26641161 -----------CCGCAGAA-GUGCGCUGAAAAGCAGACUACAGUCUGCGAUACGCACCACUUUCUGCGGUUCGCUGUGCAUAUCCGUUACUCGAACAUA--- -----------.(((((((-(((((.......((((((....))))))....)))))....)))))))(((((..((((......).))).)))))...--- ( -32.30, z-score = -3.33, R) >droSec1.super_3 5073346 96 - 7220098 ACUCGCAUC--CCGCAGAA-GUGCGCUGAAAAGCAGACUACAGAAUGCGGAACGCACCACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUA--- ....(((..--((((((((-(((.(((....)))...........((((...)))).))).))))))))..))).........................--- ( -29.80, z-score = -1.95, R) >droSim1.chr2L 9399619 96 - 22036055 ACUCGCAUC--CCGCAGAA-GUGAGCUGAAAAGCAGACUACAGAAUGCGGAACGCACCACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUA--- ....(((..--((((((((-(((.(((....)))...........((((...)))).))).))))))))..))).........................--- ( -30.40, z-score = -2.40, R) >droGri2.scaffold_15252 13654200 101 + 17193109 AUUCGUAGCAGCCACAAAAUAUGCGGCCAACUGGUGAAUGGAAAUUGUGGUGCAUGUUGUUGU-UCUUGUAGGCUAUCUUUAAUUUGCUCUCCAAAUUGUGC .......((((((((((((((((((.(((...(((........))).))))))))))).....-..)))).)))).....(((((((.....))))))).)) ( -20.31, z-score = 1.13, R) >consensus ACUCGCAUC__CCGCAGAA_GUGCGCUGAAAAGCAGACUACAGAAUGCGAAACGCACCACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUA___ ...........((((((((.(((.(((....)))...........((((...)))).))).))))))))................................. (-11.00 = -12.37 + 1.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:38 2011