| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,365,150 – 1,365,240 |
| Length | 90 |
| Max. P | 0.700470 |

| Location | 1,365,150 – 1,365,240 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 66.21 |
| Shannon entropy | 0.73542 |
| G+C content | 0.48228 |
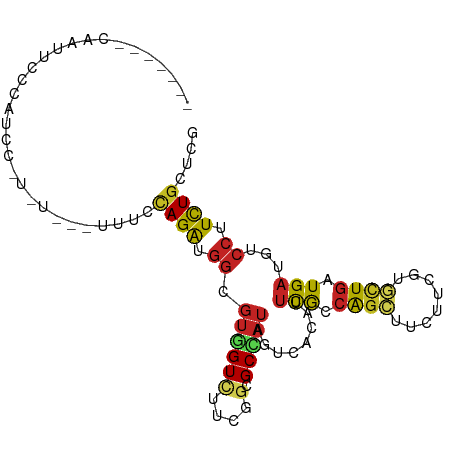
| Mean single sequence MFE | -20.51 |
| Consensus MFE | -10.16 |
| Energy contribution | -9.96 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.44 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
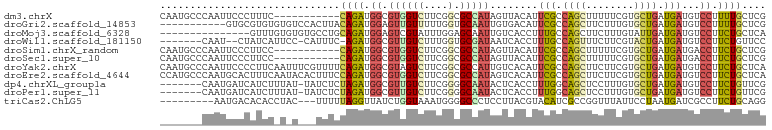
>dm3.chrX 1365150 90 + 22422827 CAAUGCCCAAUUCCCUUUC-----------CAGAUGGCGUGGUCUUCGGCGCCAUAGUUACAUUCGCCAGCUUUUUCGUGCUGAUGAUGUCCUUUUGCUCG ....((.............-----------...(((((((.(....).)))))))....((((.((.((((........)))).))))))......))... ( -21.20, z-score = -0.62, R) >droGri2.scaffold_14853 320176 90 + 10151454 -----------GUGCGUGUGUGUCCACUUACAGAUGGAGUUGUUUUUGGUGCAAUUGUGACAUUCGCCAGCUUCUUUGUGCUGAUGAUGUCCUUUUGCUCG -----------...(((.((((......)))).)))............(.((((..(.(((((.((.((((........)))).))))))))..)))).). ( -24.20, z-score = -1.08, R) >droMoj3.scaffold_6328 441684 86 - 4453435 ---------------GUUUGUGUGUGCCUGCAGAUGGAGUCGUAUUUGGAGCAAUUGUCACCUUUGCCAGCUUCUUUGUAUUGAUGAUGUCCUUCUGCUCA ---------------..............(((((.((((((((((..(((((...((.((....)).)))))))...))).))))....))).)))))... ( -18.10, z-score = 0.48, R) >droWil1.scaffold_181150 3682774 90 - 4952429 -------CAAU--CUAUCAUUCC-CAUUUC-AGAUGGCGUUGUCUUUGGUGCGAUAAUCACCUUUGCCAGUUUCUUCGUACUGAUGAUGUCCUUCUGUUCC -------....--..........-.....(-(((.(((((..((...(((((((.....((........))....)))))))))..)))))..)))).... ( -19.80, z-score = -2.06, R) >droSim1.chrX_random 988721 90 + 5698898 CAAUGCCCAAUUCCCUUCC-----------CAGAUGGCGUGGUCUUCGGCGCCAUAGUUACAUUCGCCAGCUUUUUCGUGCUGAUGAUGACCUUCUGCUCG ...................-----------((((((((((.(....).))))))..((((...(((.((((........)))).)))))))..)))).... ( -22.00, z-score = -0.65, R) >droSec1.super_10 1192136 90 + 3036183 CAAUGCCCAAUUCCCUUCC-----------CAGAUGGCGUGGUCUUCGGCGCCAUAGUUACAUUCGCCAGCUUUUUCGUGCUGAUGAUGACCUUCUGCUCG ...................-----------((((((((((.(....).))))))..((((...(((.((((........)))).)))))))..)))).... ( -22.00, z-score = -0.65, R) >droYak2.chrX 1269862 101 + 21770863 CAAUGCCCAAUUCCCCUUCAAUUUCGUUUUCAGAUGGCGUAGUCUUCGGCGCCAUUGUCACAUUCGCCAGCUUCUUCGUGCUGAUGAUGUCCUUCUGCUCA ....((..........................((((((((........))))))))...((((.((.((((........)))).))))))......))... ( -20.60, z-score = -0.55, R) >droEre2.scaffold_4644 1354639 101 + 2521924 CCAUGCCCAAUGCACUUUCAAUACACUUUCCAGAUGGCGUGGUCUUCGGCGCCAUAGUCACAUUCGCCAGCUUCUUCGUGCUGAUGAUGUCCUUCUGCUCA ...(((.....)))................((((((((((.(....).)))))).....((((.((.((((........)))).))))))...)))).... ( -23.20, z-score = -0.11, R) >dp4.chrXL_group1a 4498834 93 - 9151740 -------CAAUGAUCAUCUUUAU-UAUCUCUAGAUGGCGUUGUCUUCGGGGCAAUACUCACCUUUGGCAGCUCCUUUGUGCUGAUGAUGUCCUUCUGUUCG -------(((((.((((((....-.......))))))))))).....((((((.......((...))((((........))))....))))))........ ( -19.70, z-score = -0.38, R) >droPer1.super_11 1605236 93 - 2846995 -------CAAUGAUCAUCUUUAU-UAUCUCUAGAUGGCGUUGUCUUCGGGGCAAUACUCACCUUUGGCAGCUCCUUUGUGCUGAUGAUGUCCUUCUGUUCG -------(((((.((((((....-.......))))))))))).....((((((.......((...))((((........))))....))))))........ ( -19.70, z-score = -0.38, R) >triCas2.ChLG5 11770401 89 + 18847211 ---------AAUGACACACCUAC---UUUUUAGGUUAUCUGGUAAAUGGGGCCCUCCUUACGUACAUCGCCGGUUUAUUCCUAAUGAUCGCCUUCUGCAGG ---------.........(((.(---.....((((.(((..(((..(((((....)))))..)))......((......))....))).))))...).))) ( -15.10, z-score = 1.19, R) >consensus _______CAAUUCCCAUCC_U_U___UUUCCAGAUGGCGUGGUCUUCGGCGCCAUAGUCACAUUCGCCAGCUUCUUCGUGCUGAUGAUGUCCUUCUGCUCG ..............................((((.((....(((.(((((((.........................))))))).)))..)).)))).... (-10.16 = -9.96 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:06:05 2011