| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,267,077 – 1,267,175 |
| Length | 98 |
| Max. P | 0.580709 |

| Location | 1,267,077 – 1,267,175 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.19 |
| Shannon entropy | 0.19189 |
| G+C content | 0.30198 |
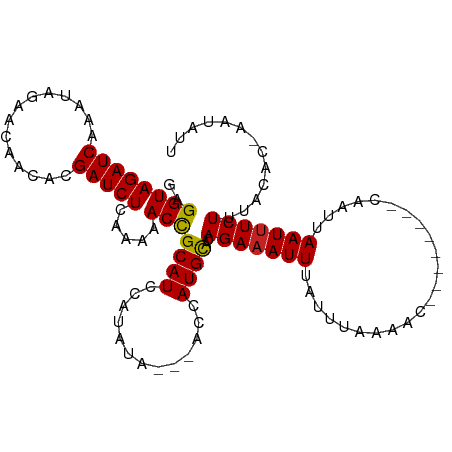
| Mean single sequence MFE | -10.43 |
| Consensus MFE | -7.81 |
| Energy contribution | -7.67 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
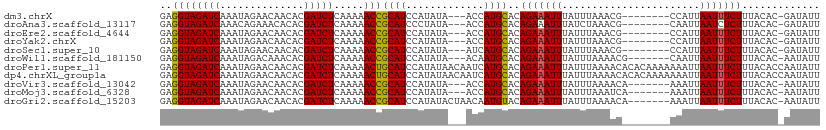
>dm3.chrX 1267077 98 + 22422827 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACCGCAUCCAUAUA---ACCAUGCACAGAAAUUUAUUUAAACG--------CCAUUAAUUUCUUUACAC-GAUAUU ..((((((((..............))))).....)))((((.......---...))))..(((((((..........--------.....)))))))......-...... ( -9.30, z-score = -1.08, R) >droAna3.scaffold_13117 3932367 98 - 5790199 GAGGUAGAUCAAACAGAAACACACGAUCUCAAAAACCGCAUCCCUAUA---ACCAUGCACAGAAAUUUAUCUAAACG--------CAAUUAAUCUCUUUACAC-GAUAUU ((((((((((..............)))))........((((.......---...))))...................--------......))))).......-...... ( -9.14, z-score = -0.93, R) >droEre2.scaffold_4644 1250769 98 + 2521924 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACCGCAUCCAUAUA---ACCAUGCACAGAAAUUUAUUUAAACG--------CCAUUAAUUUCUUUACAC-GAUAUU ..((((((((..............))))).....)))((((.......---...))))..(((((((..........--------.....)))))))......-...... ( -9.30, z-score = -1.08, R) >droYak2.chrX 1174775 98 + 21770863 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACCGCAUCCAUAUA---ACCAUGCACAGAAAUUUAUUUAAACG--------CCAUUAAUUUCUUUACAC-GAUAUU ..((((((((..............))))).....)))((((.......---...))))..(((((((..........--------.....)))))))......-...... ( -9.30, z-score = -1.08, R) >droSec1.super_10 1090514 98 + 3036183 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACCGCAUCCAUAUA---AUCAUGCACAGAAAUUUAUUUAAACG--------CCAUUAAUUUCUUUACAC-GAUAUU ..((((((((..............))))).....)))...........---(((.((...(((((((..........--------.....)))))))...)).-)))... ( -9.50, z-score = -0.89, R) >droWil1.scaffold_181150 418157 99 + 4952429 GAGGUAGAUCAAAUAGACAAACACGAUCUCAAAAACCGCAUCCAUAUA---ACAAUGCACAGAAAUUUAUUUAAAACG-------CAAUUAAUUUCUUUACAC-AAUAUU ..((((((((..............))))).....)))((((.......---...))))..(((((((.(((.......-------.))).)))))))......-...... ( -10.24, z-score = -1.92, R) >droPer1.super_11 1476775 110 - 2846995 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACUGCAUCCAUAUAACAAUCAUGCACAGAAAUUUAUUUAAAACACACAAAAAAAUUAAUUUCUUUACACCAAUAUU ..((((((((..............))))).......(((((.............))))).(((((((.((((.............)))).)))))))....)))...... ( -10.48, z-score = -2.05, R) >dp4.chrXL_group1a 4368575 110 - 9151740 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACUGCAUCCAUAUAACAAUCAUGCACAGAAAUUUAUUUAAAACACACAAAAAAAUUAAUUUCUUUACACCAAUAUU ..((((((((..............))))).......(((((.............))))).(((((((.((((.............)))).)))))))....)))...... ( -10.48, z-score = -2.05, R) >droVir3.scaffold_13042 2573916 99 - 5191987 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACCGCAUCCAUAUA---ACCAUGCACAGAAAUUUAUUUAAAACA-------AAAUUAAUUUCUUUACAC-AAUAUU ..((((((((..............))))).....)))((((.......---...))))..(((((((.((((......-------)))).)))))))......-...... ( -11.64, z-score = -3.04, R) >droMoj3.scaffold_6328 978079 99 + 4453435 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACCGCAUCCAUAUA---ACCAUGCACAGAAAUUUAUUUAAAUCA-------AAAUUAAUUUCUUUACAC-AAUAUU ..((((((((..............))))).....)))((((.......---...))))..(((((((.((((......-------)))).)))))))......-...... ( -11.64, z-score = -2.81, R) >droGri2.scaffold_15203 6557641 102 - 11997470 GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACCGCAUCCAUAUACUAACAAUGUACAGAAAUUUAUUUAAAACA-------AAAUUAAUUUCUUUACAC-AAUAUU ..((((((((..............))))).....)))..................((((.(((((((.((((......-------)))).))))))).)))).-...... ( -13.74, z-score = -3.46, R) >consensus GAGGUAGAUCAAAUAGAACAACACGAUCUCAAAAACCGCAUCCAUAUA___ACCAUGCACAGAAAUUUAUUUAAAAC________CAAUUAAUUUCUUUACAC_AAUAUU ..((((((((..............))))).....)))((((.............))))..(((((((.......................)))))))............. ( -7.81 = -7.67 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:55 2011