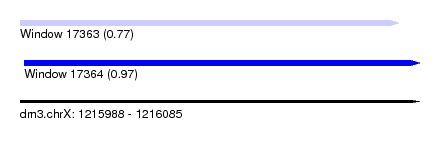
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,215,988 – 1,216,085 |
| Length | 97 |
| Max. P | 0.969260 |

| Location | 1,215,988 – 1,216,080 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 56.83 |
| Shannon entropy | 0.58258 |
| G+C content | 0.48917 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -4.48 |
| Energy contribution | -5.60 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1215988 92 + 22422827 ACAC-AAAAGUUGCAUUAAUCGCAACC---CCGGUAAAACGGGAAUUGCUUGCAGGCUCUCUGCGCCUCUUUCUCUCUCUCUCACUCCGUCCCACU ....-.((((..((.......((((.(---(((......))))..))))..((((.....))))))..))))........................ ( -20.10, z-score = -2.09, R) >droYak2.chrX 1133770 82 + 21770863 ACAC-AAUAAGUGCAUUAAUCGCAACC---CCGGUAAAACGGCA----UUUGCAGGCUCUCUGCGCCUCUCUCUCCUUCU----CUCUGUCGCA-- ....-.....(((((......((((..---(((......)))..----.))))((((.......))))............----...)).))).-- ( -15.40, z-score = -0.80, R) >droGri2.scaffold_15081 1802308 90 - 4274704 ACAAGAACAACUGCAAUAGUUGUUGUUGUUCUUGUUGAGCCGCUGU--UAAGCGGUCCCCCUCCCUUUAUUUUUCUUCCCUU--UUGCAACCCA-- (((((((((((.(((.....))).))))))))))).(..(((((..--..)))))..)........................--..........-- ( -25.60, z-score = -5.39, R) >consensus ACAC_AAAAACUGCAUUAAUCGCAACC___CCGGUAAAACGGCA_U__UUUGCAGGCUCUCUGCGCCUCUUUCUCUUUCU_U__CUCCGUCCCA__ .........(((((.......)))))....(((......))).........((((.....))))................................ ( -4.48 = -5.60 + 1.12)

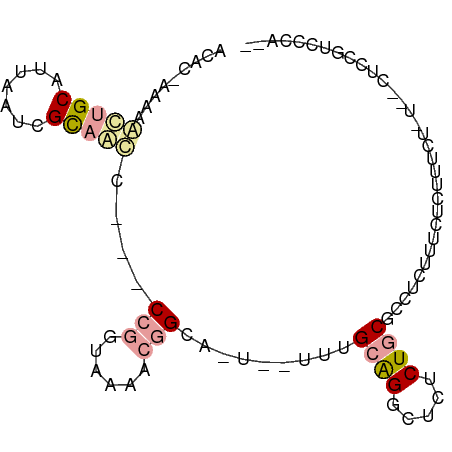
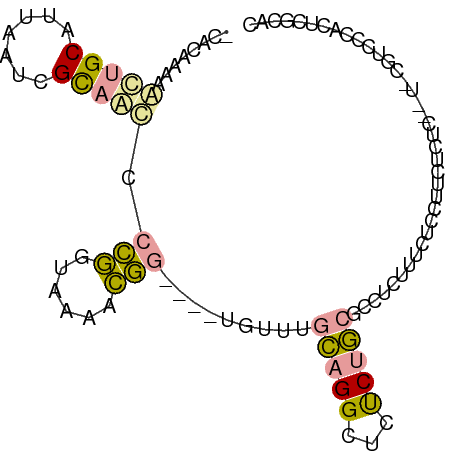
| Location | 1,215,989 – 1,216,085 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 53.63 |
| Shannon entropy | 0.65140 |
| G+C content | 0.51163 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -5.31 |
| Energy contribution | -5.10 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1215989 96 + 22422827 -CACAAAAGUUGCAUUAAUCGCAACCCCGGUAAAACGGGAAUUGCUUGCAGGCUCUCUGCGCCUCUUUCUCUCUCUCUCACUCCGUCCCACUCUCAC -....((((..((.......((((.((((......))))..))))..((((.....))))))..))))............................. ( -20.10, z-score = -2.20, R) >droYak2.chrX 1133771 86 + 21770863 -CACAAUAAGUGCAUUAAUCGCAACCCCGGUAAAACGG----CAUUUGCAGGCUCUCUGCGCCUCUCUCUCCUUCUCUC----UGUCGCACUCGC-- -.......(((((.......((((..(((......)))----...))))((((.......))))...............----....)))))...-- ( -19.40, z-score = -2.09, R) >droGri2.scaffold_15081 1802309 96 - 4274704 CAAGAACAACUGCAAUAGUUGUUGUUGUUCUUGUUGAGCCGCUGUUAAGCGGUCCCCCUCCCUUUAUUUUUCUUCCCUU-UUGCAACCCACCCGCAC ((((((((((.(((.....))).))))))))))..(..(((((....)))))..)........................-.(((.........))). ( -25.20, z-score = -4.40, R) >consensus _CACAAAAACUGCAUUAAUCGCAACCCCGGUAAAACGG____UGUUUGCAGGCUCUCUGCGCCUCUUUCUCCUUCUCUC__U_CGUCCCACUCGCAC ........(((((.......))))).(((......))).........(((((...)))))..................................... ( -5.31 = -5.10 + -0.21)

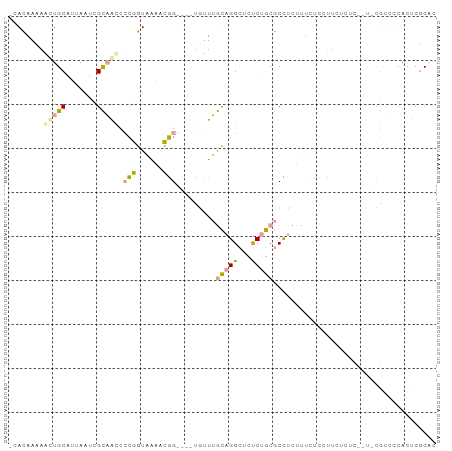
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:51 2011