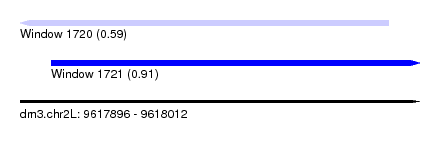
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,617,896 – 9,618,012 |
| Length | 116 |
| Max. P | 0.908957 |

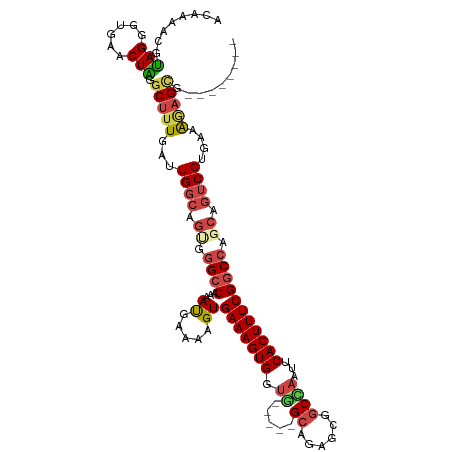
| Location | 9,617,896 – 9,618,003 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.16 |
| Shannon entropy | 0.37262 |
| G+C content | 0.51452 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9617896 107 - 23011544 ACAAAACGUAGGGUGAACUAGGCUUUGAUUGGCAGUGGGCCAAAAUGAAAAGUGAAAGUGGUG-----GCAGAGUGGCCAAUUCACUUUCGGCCAGCAGUCGUGAAAGAGCG-------- ........(((......))).(((((...((((.((.((((....(....)..((((((((((-----((......))))..)))))))))))).)).))))....))))).-------- ( -33.50, z-score = -1.78, R) >droYak2.chr2L 12302826 115 - 22324452 ACAAAUCGUAGGGUGACCUGAGCUUUGAUUGGCAGCGGGCCAAAACGGAAAGUGAAAGUGGUU-----GCAGAGCGGCCAAUUCACUUUCGGCCAACAGUCGAAAGGCAGUGCAAAAGCA ........((((....)))).(((((.....((((..((((......(((((((((..(((((-----((...))))))).)))))))))))))..).(((....)))..))).))))). ( -46.80, z-score = -3.89, R) >droEre2.scaffold_4929 10225121 112 - 26641161 ACAAAUCGUAGGGUGACCUGGGCUUUGAUUGGCAGUGGGCCAAAACGGAAAGUGAAAGUGGUGGCAGAGCAGAGCGGCCAAUUCACUUUCGGCCAGCAGUCGCGUAGAAGCA-------- .((((.(.((((....)))).).))))...(((.((.((((......(((((((((..((((.((........)).)))).))))))))))))).)).)))((......)).-------- ( -44.40, z-score = -2.90, R) >droSec1.super_3 5064216 102 - 7220098 ACAAAACGUAGGGUGAACUAGGCUUUGAUUGGCAGUGGGCCAAAAUGAAAAGUGAAAGUGCUG-----GCAGAGCGACUAAUUCACUUUCGGCCG-----CGUGAAAGAGCG-------- ......((.((((((((.(((((((((.(((((.....))))).......(((......))).-----.))))))..))).))))))))))..((-----(........)))-------- ( -27.90, z-score = -0.44, R) >droSim1.chrU 11836270 107 - 15797150 ACAAAACGUAGGGUGAACUAGGCUUUGAUUGGCAGUGGGCCAAAAUGAAAAGUGAAAGUGCUG-----GCAGAGCGGCCAAUUCACUUUCGGCCAGCAGUCGUGAAAGAGCG-------- ........(((......))).(((((...((((.((.((((....(....)..(((((((.((-----((......))))...))))))))))).)).))))....))))).-------- ( -32.40, z-score = -0.80, R) >droAna3.scaffold_12943 1044138 102 - 5039921 UCAGAGUACAGGGUA--CUGCUCUUU--CUGGCCGCGCGCC-AAAUGAAAAGUGAAAGUGGUC-----GCAGAGCGGCGUACUCACUUUCGACCAGCAGUCGUGAGAAUGCA-------- ...((((((......--(((((((..--.(((((((.(((.-.........)))...))))))-----).))))))).))))))...........(((.((....)).))).-------- ( -35.00, z-score = -0.96, R) >consensus ACAAAACGUAGGGUGAACUAGGCUUUGAUUGGCAGUGGGCCAAAAUGAAAAGUGAAAGUGGUG_____GCAGAGCGGCCAAUUCACUUUCGGCCAGCAGUCGUGAAAGAGCG________ ........(((......))).(((((...((((.((.((((...((.....))((((((((.......((...)).......)))))))))))).)).))))....)))))......... (-19.99 = -20.66 + 0.67)

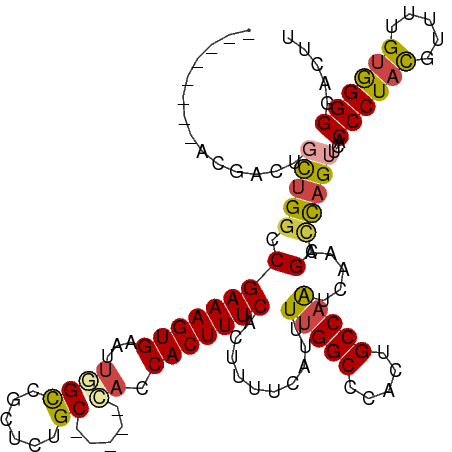
| Location | 9,617,905 – 9,618,012 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Shannon entropy | 0.33750 |
| G+C content | 0.52365 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -23.15 |
| Energy contribution | -22.88 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9617905 107 + 23011544 --------ACGACUGCUGGCCGAAAGUGAAUUGGCCACUCUGC-----CACCACUUUCACUUUUCAUUUUGGCCCACUGCCAAUCAAAGCCUAGUUCACCCUACGUUUUGUGGGGGACUU --------..((..((((((.(((((((...((((......))-----)).)))))))..........(((((.....))))).....))).))))).((((((.....))))))..... ( -34.40, z-score = -2.66, R) >droYak2.chr2L 12302835 115 + 22324452 ACUGCCUUUCGACUGUUGGCCGAAAGUGAAUUGGCCGCUCUGC-----AACCACUUUCACUUUCCGUUUUGGCCCGCUGCCAAUCAAAGCUCAGGUCACCCUACGAUUUGUGGGGCACUU ..........((((...(((.(((((((((.(((..((...))-----..)))..)))))))))....(((((.....))))).....)))..)))).((((((.....))))))..... ( -35.30, z-score = -1.29, R) >droEre2.scaffold_4929 10225130 112 + 26641161 --------GCGACUGCUGGCCGAAAGUGAAUUGGCCGCUCUGCUCUGCCACCACUUUCACUUUCCGUUUUGGCCCACUGCCAAUCAAAGCCCAGGUCACCCUACGAUUUGUAGGGGACUU --------..((((...(((.(((((((((.(((..((........))..)))..)))))))))....(((((.....))))).....)))..)))).((((((.....))))))..... ( -40.70, z-score = -3.07, R) >droSec1.super_3 5064225 102 + 7220098 ------------ACGC-GGCCGAAAGUGAAUUAGUCGCUCUGC-----CAGCACUUUCACUUUUCAUUUUGGCCCACUGCCAAUCAAAGCCUAGUUCACCCUACGUUUUGUGGGGGACUU ------------....-(((.(((((((((..(((.(((....-----.)))))))))))))))....(((((.....))))).....))).(((.(.((((((.....)))))))))). ( -30.90, z-score = -1.88, R) >droSim1.chrU 11836279 107 + 15797150 --------ACGACUGCUGGCCGAAAGUGAAUUGGCCGCUCUGC-----CAGCACUUUCACUUUUCAUUUUGGCCCACUGCCAAUCAAAGCCUAGUUCACCCUACGUUUUGUGGGGGACUU --------..((..((((((.(((((((...((((......))-----)).)))))))..........(((((.....))))).....))).))))).((((((.....))))))..... ( -35.50, z-score = -2.18, R) >droAna3.scaffold_12943 1044147 102 + 5039921 --------ACGACUGCUGGUCGAAAGUGAGUACGCCGCUCUGC-----GACCACUUUCACUUUUCAUUU-GGCGCGCGGCCAG--AAAGAGCAGU--ACCCUGUACUCUGAGGGGUAUUU --------...(((((((((.(((((((.((.(((......))-----))))))))))))).....(((-(((.....)))))--)...))))))--(((((.........))))).... ( -36.00, z-score = -1.64, R) >consensus ________ACGACUGCUGGCCGAAAGUGAAUUGGCCGCUCUGC_____CACCACUUUCACUUUUCAUUUUGGCCCACUGCCAAUCAAAGCCCAGUUCACCCUACGUUUUGUGGGGGACUU ..............(((((.((((((((((.........................)))))))))....(((((.....))))).....).)))))...((((((.....))))))..... (-23.15 = -22.88 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:35 2011