
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,197,316 – 1,197,414 |
| Length | 98 |
| Max. P | 0.630914 |

| Location | 1,197,316 – 1,197,414 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.30 |
| Shannon entropy | 0.54760 |
| G+C content | 0.40509 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -17.08 |
| Energy contribution | -14.01 |
| Covariance contribution | -3.07 |
| Combinations/Pair | 1.91 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
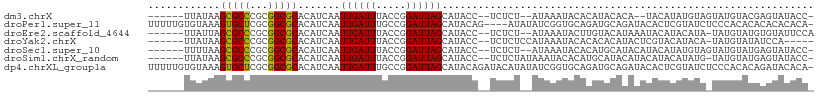
>dm3.chrX 1197316 98 + 22422827 -GGUAUACUCGUACAUACUACAUAUGUA--UGUGUAUGUGUAUUUAU--AGAGA--GGUAUGCUAAUCCGGUAAAUCAAUUGAUGUGCGCCGCGGGCGCUUAUAA------ -...((((.((((((((.(((....)))--)))))))).))))((((--((...--(((.((((.....)))).))).........(((((...)))))))))))------ ( -24.60, z-score = 0.27, R) >droPer1.super_11 1400506 106 - 2846995 -UGUGUGUGUGUGUGGGAGAUACGAGUGUAUCUGCAUCUGCACCGAUAUAU----CUGUAUGCUAAUCCGGCAAAUCAAUUGAUGUGCGCCGCGAGCACUUUACACAAAAA -(((((((((.(((((.(((((((..(((((((((....)))..)))))).----.))))).))......(((.(((....))).))).))))).))))...))))).... ( -33.70, z-score = -1.64, R) >droEre2.scaffold_4644 1188916 100 + 2521924 UGGAAUACACAUACAUA-UAUGUAUGUAUUUAUGUACAAGUAUUUAU--AGAGA--GGUAUGCUAAUCCGGUAAAUCAAUUGAUGUGCGCCGCGGGCGCUAAUAA------ ..(((((((((((....-))))).(((((....))))).))))))..--.....--(((.((((.....)))).))).........(((((...)))))......------ ( -20.60, z-score = 0.64, R) >droYak2.chrX 1113704 97 + 21770863 -----UGGAUAUACAUA-UGUAUGUACGAGUAUGUGUGUGUAUUUAUGGAGAGA--GGUAUGCUAAUCCGGUAAAUCAAUUGAUGUGCGCCGCGGGCGCUUAUAA------ -----((((((((((((-(((((......)))))))))))))))))((((...(--(.....))..))))............(((.(((((...))))).)))..------ ( -27.50, z-score = -1.11, R) >droSec1.super_10 1030450 100 + 3036183 -GGUAUACUCAUACAUACUACAUAUGUAUGUAUGCAUGUGUAUUUAU--AGAGA--GGUAUGCUAAUCCGGUAAAUCAAUUGAUGUGCGCCGCGGGCGCUUAAAA------ -(((((((.(((((((((.......)))))))))...)))))))...--.....--(((.((((.....)))).))).........(((((...)))))......------ ( -26.30, z-score = -0.51, R) >droSim1.chrX_random 937985 101 + 5698898 -GGUAUACUCAUACAUA-CAUAUGUAUGUAUGUAUGCAUGUGUAUUUAUAGAGA--GGUAUGCUAAUCCGGUAAAUCAAUUGAUGUGCGCCGCGGGCGCUUAUAA------ -......((((((.(((-((((((((((....))))))))))))).))).))).--(((.((((.....)))).))).....(((.(((((...))))).)))..------ ( -29.60, z-score = -0.95, R) >dp4.chrXL_group1a 4290698 110 - 9151740 -UGUGUAUCUGUGUGGGAGAUACGAGUGUAUCUGCAUCUGCACCGAUAUAUGUAUCUGUAUGCUAAUCCGGCAAAUCAAUUGAUGUGCGCCGCGAGCACUUUACACAAAAA -((((((...((((.(.((((((..((((((((((....)))..))))))))))))).).......((((((..(((....)))....)))).))))))..)))))).... ( -33.80, z-score = -1.78, R) >consensus _GGUAUACUCAUACAUA_UAUAUGUGUGUAUAUGUAUGUGUAUUUAU__AGAGA__GGUAUGCUAAUCCGGUAAAUCAAUUGAUGUGCGCCGCGGGCGCUUAUAA______ ..........(((((((.(((((....)))))....))))))).................((((....((((..(((....)))....))))..))))............. (-17.08 = -14.01 + -3.07)

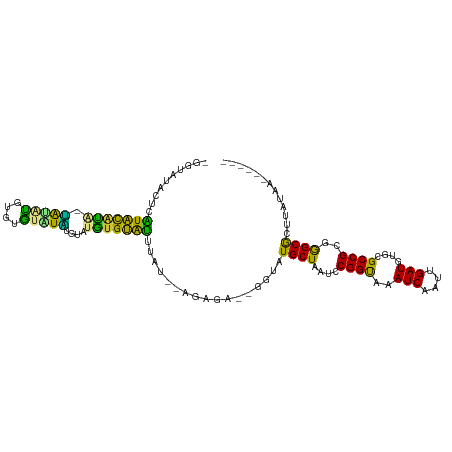
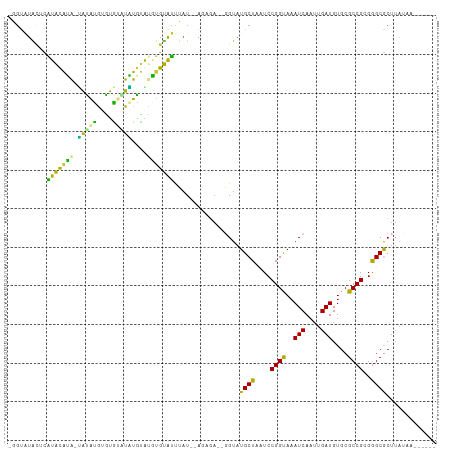
| Location | 1,197,316 – 1,197,414 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.30 |
| Shannon entropy | 0.54760 |
| G+C content | 0.40509 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.27 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1197316 98 - 22422827 ------UUAUAAGCGCCCGCGGCGCACAUCAAUUGAUUUACCGGAUUAGCAUACC--UCUCU--AUAAAUACACAUACACA--UACAUAUGUAGUAUGUACGAGUAUACC- ------(((((.(((((...))))).......((((((.....))))))......--....)--))))(((((((((((((--(....)))).))))))....))))...- ( -18.50, z-score = -0.81, R) >droPer1.super_11 1400506 106 + 2846995 UUUUUGUGUAAAGUGCUCGCGGCGCACAUCAAUUGAUUUGCCGGAUUAGCAUACAG----AUAUAUCGGUGCAGAUGCAGAUACACUCGUAUCUCCCACACACACACACA- ....(((((....(((((.(((((...(((....))).))))).....((((...(----(....)).)))).)).)))(((((....)))))....)))))........- ( -25.30, z-score = -0.66, R) >droEre2.scaffold_4644 1188916 100 - 2521924 ------UUAUUAGCGCCCGCGGCGCACAUCAAUUGAUUUACCGGAUUAGCAUACC--UCUCU--AUAAAUACUUGUACAUAAAUACAUACAUA-UAUGUAUGUGUAUUCCA ------......(((((...))))).................(((..........--....(--((((....))))).....(((((((((..-..)))))))))..))). ( -20.30, z-score = -1.44, R) >droYak2.chrX 1113704 97 - 21770863 ------UUAUAAGCGCCCGCGGCGCACAUCAAUUGAUUUACCGGAUUAGCAUACC--UCUCUCCAUAAAUACACACACAUACUCGUACAUACA-UAUGUAUAUCCA----- ------......(((((...)))))........(((((((..(((..((.....)--)...))).))))).))...........((((((...-.)))))).....----- ( -14.00, z-score = -0.82, R) >droSec1.super_10 1030450 100 - 3036183 ------UUUUAAGCGCCCGCGGCGCACAUCAAUUGAUUUACCGGAUUAGCAUACC--UCUCU--AUAAAUACACAUGCAUACAUACAUAUGUAGUAUGUAUGAGUAUACC- ------......(((((...))))).......((((((.....))))))......--.....--....((((.(((((((((.(((....)))))))))))).))))...- ( -25.40, z-score = -2.62, R) >droSim1.chrX_random 937985 101 - 5698898 ------UUAUAAGCGCCCGCGGCGCACAUCAAUUGAUUUACCGGAUUAGCAUACC--UCUCUAUAAAUACACAUGCAUACAUACAUACAUAUG-UAUGUAUGAGUAUACC- ------(((((.(((((...))))).......((((((.....))))))......--....)))))((((.((((((((((((......))))-)))))))).))))...- ( -27.30, z-score = -3.42, R) >dp4.chrXL_group1a 4290698 110 + 9151740 UUUUUGUGUAAAGUGCUCGCGGCGCACAUCAAUUGAUUUGCCGGAUUAGCAUACAGAUACAUAUAUCGGUGCAGAUGCAGAUACACUCGUAUCUCCCACACAGAUACACA- ..(((((((....(((((.(((((...(((....))).))))).....((((...((((....)))).)))).)).)))(((((....)))))....)))))))......- ( -29.50, z-score = -1.80, R) >consensus ______UUAUAAGCGCCCGCGGCGCACAUCAAUUGAUUUACCGGAUUAGCAUACC__UCUCU__AUAAAUACACAUACAUAUACACACAUAUA_UAUGUAUGAGUAUACC_ ............(((((...))))).......((((((.....)))))).............................................................. ( -8.68 = -8.27 + -0.41)

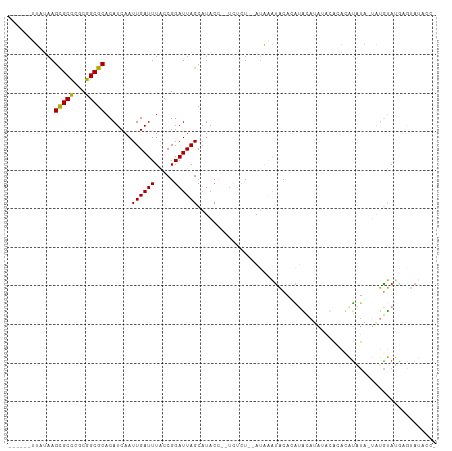
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:43 2011