
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,147,046 – 1,147,294 |
| Length | 248 |
| Max. P | 0.808226 |

| Location | 1,147,046 – 1,147,166 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 65.62 |
| Shannon entropy | 0.46907 |
| G+C content | 0.51028 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.63 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
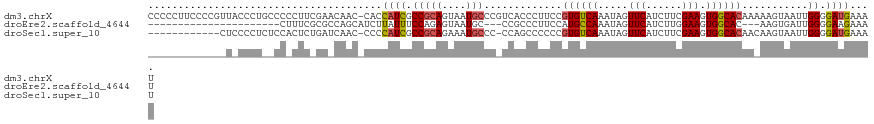
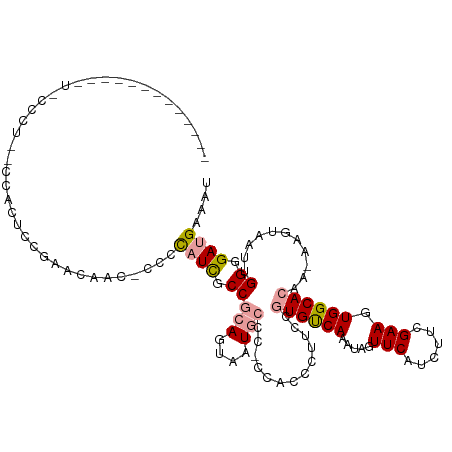
>dm3.chrX 1147046 120 - 22422827 CCCCCUUCCCCGUUACCCUGCCCCCUUCGAACAAC-CACCAUCGCCGCAGUAAUGCCCGUCACCCUUCCGUGUCAAAUAGUUCAUCUUCGAAGUGGCACAAAAAGUAAUUGGGGAUGAAAU ......((((((((((..((((..(((((((....-..........(((....)))..(.(((......))).)............))))))).))))......)))).))))))...... ( -30.30, z-score = -3.15, R) >droEre2.scaffold_4644 1133825 93 - 2521924 ----------------------CUUUCGCGCCAGCAUCUUAUUUCCAGAGUAAUGC---CCGCCCUUCCAUGCCAAAUAGUUCAUCUUGGAAGUGGCAC---AAGUGAUUGGGGAAGAAAU ----------------------.((((((....))....................(---(((((((((((.................)))))).))).(---((....))))))..)))). ( -23.53, z-score = -0.47, R) >droSec1.super_10 980636 107 - 3036183 ------------CUCCCCUCUCCACUCUGAUCAAC-CCCCAUCGCCGCAGAAAUGCCC-CCAGCCCCCCGUGUCAAAUAGUUCAUCUUCGAAGUGGCACAACAAGUAAUUGGGGAUGAAAU ------------..................(((.(-(((.((..(.(((....)))..-..........((((((....(........)....)))))).....)..)).)))).)))... ( -23.40, z-score = -1.23, R) >consensus _____________U_CCCU__CCACUCCGAACAAC_CCCCAUCGCCGCAGUAAUGCCC_CCACCCUUCCGUGUCAAAUAGUUCAUCUUCGAAGUGGCACAA_AAGUAAUUGGGGAUGAAAU .......................................((((.(((((....))).............((((((.....(((......))).))))))...........)).)))).... (-13.96 = -14.63 + 0.67)
| Location | 1,147,166 – 1,147,294 |
|---|---|
| Length | 128 |
| Sequences | 4 |
| Columns | 138 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Shannon entropy | 0.30366 |
| G+C content | 0.46848 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -25.95 |
| Energy contribution | -28.20 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
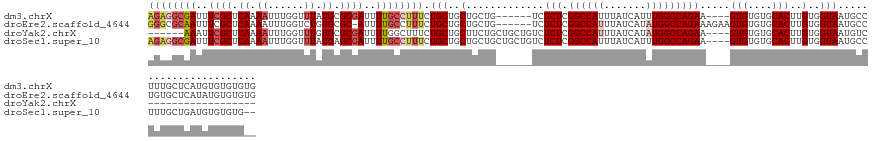
>dm3.chrX 1147166 128 + 22422827 AGAGGCGAUUUCGCUCAAAAUUUGGUUUAUGCGCGAUUUUGCCUUUCUGCUGCUGCUG------UCUCUCGGCCAUUUAUCAUUUGGCCAGAA----GUGUGUGCACUUGUGGUAAUGCCUUUGCUCAUGUGUGUGUG ((((((((..((((.((.((......)).)).))))..))))))))..((....))..------..(((.(((((.........)))))))).----....(..(((.((((((((.....)))).)))).)))..). ( -38.30, z-score = -1.34, R) >droEre2.scaffold_4644 1133918 131 + 2521924 GGGCGCAAUUUCGCUCAAAAUUUGGUCUGUGCGC-AUUUUGCCUUUCUGCUGCUGCUG------UCUCUCGGCCAUUUAUCAUAUGGCCAGAAAGAAGUGUGUGCACUUGUGGUAAUGCCUGUGCUCAUAUGUGUGUG (((((......)))))..............((((-((.....((((((((....))..------......((((((.......))))))))))))..(((((.((((....((.....)).)))).))))))))))). ( -40.90, z-score = -1.38, R) >droYak2.chrX 1061712 110 + 21770863 ------AAAUUCGCUCAAAAUUUGGUUUGUGCGCGAUUUUGGCUUUCUGCUGCUUCUGCUGCUGUCUCUCGGCCAUUUAUCAUAUGGCCAGAA----GUGUGUGCACUUGUGGUAAUGUC------------------ ------.....(((.((((......)))).))).(((..(((((..(.((.((....)).)).)......)))))...)))((((.((((.((----(((....))))).)))).)))).------------------ ( -31.50, z-score = -1.85, R) >droSec1.super_10 980743 132 + 3036183 AGAGGCGAUUUCGCUCAAAAUUUGGUUUAUGAGCGAUUUUGCCUUUCUGCUGCUGCUGCUGCUGUCUCUCGGCCAUUUAUCAUUUGGCCAGAA----GUGUGUGCACUUGUGGUAAUGCCUUUGCUGAUGUGUGUG-- ((((((((..(((((((.((......)).)))))))..))))))))..((.((....)).))....(((.(((((.........)))))))).----....(..(((...((((((.....))))))..)))..).-- ( -44.80, z-score = -2.74, R) >consensus AGAGGCAAUUUCGCUCAAAAUUUGGUUUAUGCGCGAUUUUGCCUUUCUGCUGCUGCUG______UCUCUCGGCCAUUUAUCAUAUGGCCAGAA____GUGUGUGCACUUGUGGUAAUGCCUUUGCUCAUGUGUGUG__ ((((((((..((((.((.((......)).)).))))..)))))))).(((..(.............(((.((((((.......))))))))).....(((....)))..)..)))....................... (-25.95 = -28.20 + 2.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:33 2011