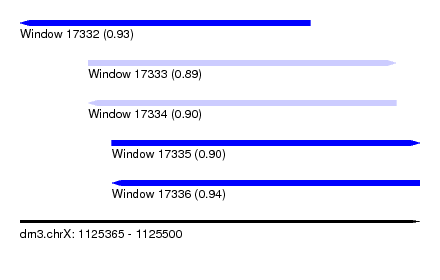
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,125,365 – 1,125,500 |
| Length | 135 |
| Max. P | 0.936588 |

| Location | 1,125,365 – 1,125,463 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 70.26 |
| Shannon entropy | 0.57537 |
| G+C content | 0.54061 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -15.85 |
| Energy contribution | -16.78 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1125365 98 - 22422827 UAUGGCAGUGCGGGCAGCGCCCUUCUGGGUGGGAAGGGA-ACUACAGGGAGAUGUGCCCGCAUCAGAUGGGCAGAUGA--AUUCAUAGAUUGGGUUAAAAU ..((.(.((((((((((..(((((((.....))))))).-.))(((......)))))))))))......).))....(--(((((.....))))))..... ( -31.30, z-score = -1.05, R) >droEre2.scaffold_4644 1110685 89 - 2521924 UAUGGCUGUGCGGGCGCCCUCUUUGCGGGAGGGAGAGG------UGGGGAGAAGUGCCCGCAUC----GGGCAGAUGA--AUUCAUAGAUUGGGUUAAAAU ..((.((((((((((((.(((((..(...........)------..)))))..))))))))).)----)).))....(--(((((.....))))))..... ( -34.70, z-score = -2.76, R) >droYak2.chrX 1040148 81 - 21770863 UAUGGCAGUGCGGGC----CCCCGCUCGACGGGAGGG---------GGGUGGGGUGCCCGCAUC-----GGCAGAUGA--AUGCAUAGAUUGGGUUAAAAU ..((.(.((((((((----((((((((..(....)..---------)))))))).)))))))).-----).))....(--((.((.....)).)))..... ( -40.40, z-score = -3.71, R) >droSec1.super_10 959496 98 - 3036183 UAUGGCAGUGCGGGCAGCGCCCUUUUGGGCGGGAAGGGA-ACUGCAGGGAGAAGUGCCCGCAUCAGAUGGGCAGAUGA--AUUCAUAGAUUGGGUUAAAAU ..((.(.(((((((((.(((((....))))).(.((...-.)).).........)))))))))......).))....(--(((((.....))))))..... ( -31.60, z-score = -1.08, R) >droSim1.chrX_random 912458 98 - 5698898 UAUGGCAGUGCGGGCAGCGCCCUUCUGGGCGGGAAGGGA-ACUGCAGGGAGAAGUGCCCGCAUCAGAUGGGCAGAUGA--AUUCAUAGAUUGGGUUAAAAU ..((.(.(((((((((.(((((....))))).(.((...-.)).).........)))))))))......).))....(--(((((.....))))))..... ( -31.60, z-score = -0.72, R) >droWil1.scaffold_181096 2579614 95 - 12416693 --AGAUAGUAAGGGUAGAG----AGUGAGAGGAAAAGCACUCCCUUUGGAGGGCUGCCCGUCGCCCGCAUAUAUGAGAUUAACAAUAGAUUGAGUUAAAAU --.(.((((....(((..(----.((((..((...(((.((((....)))).))).))..)))).)...))).....)))).).................. ( -18.90, z-score = 0.04, R) >consensus UAUGGCAGUGCGGGCAGCGCCCUUCUGGGCGGGAAGGGA_ACU_CAGGGAGAAGUGCCCGCAUCAGAUGGGCAGAUGA__AUUCAUAGAUUGGGUUAAAAU ..((((.(((((((((...(((((((.....)))))))................)))))))))...........(((......))).......)))).... (-15.85 = -16.78 + 0.92)

| Location | 1,125,388 – 1,125,492 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 63.83 |
| Shannon entropy | 0.66286 |
| G+C content | 0.55802 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -16.26 |
| Energy contribution | -17.77 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.888160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1125388 104 + 22422827 AUCUGCCCAUCUGAUGCGGGCACAUCUCCCUGUAGUUCCCUUCCCACCCAGAAGGGCGCUGCCCGCACUGCCAUAAA----AUA-------CAUAGACACUUAUAGCGGUGGGGU .....((((((...((((((((((......)))(((.((((((.......)))))).))))))))))..((.((((.----...-------.........)))).)))))))).. ( -37.02, z-score = -1.94, R) >droEre2.scaffold_4644 1110708 106 + 2521924 AUCUGCCC----GAUGCGGGCACUUCUCCCCACC-UCUCCCUCCCGCAAAGAGGGC----GCCCGCACAGCCAUAAAGUACAUAUAUACCGCAUAGGCACAGAUAGCGGUGGGGU ...(((((----(...)))))).....(((((((-...(((((.......))))).----((.......(((.....(((......)))......))).......))))))))). ( -35.04, z-score = -0.58, R) >droYak2.chrX 1040171 94 + 21770863 AUCUGCC-----GAUGCGGGCACCCCACCCC--------CCUCCCGUCGAGCGGGG----GCCCGCACUGCCAUAAA----AUAUACACCGCAUAGACACAUAAAGCUGUGGGGU ....((.-----(.(((((((.((((.(.(.--------.(....)..).).))))----)))))))).))......----......(((.(((((.(.......)))))).))) ( -33.90, z-score = -0.85, R) >droSec1.super_10 959519 104 + 3036183 AUCUGCCCAUCUGAUGCGGGCACUUCUCCCUGCAGUUCCCUUCCCGCCCAAAAGGGCGCUGCCCGCACUGCCAUAAA----AUA-------UAUAGACACUUAUAGCGGUGGGGU .....((((((...((((((((((.(.....).)))........(((((....)))))..)))))))..((.((((.----...-------.........)))).)))))))).. ( -34.82, z-score = -1.16, R) >droSim1.chrX_random 912481 104 + 5698898 AUCUGCCCAUCUGAUGCGGGCACUUCUCCCUGCAGUUCCCUUCCCGCCCAGAAGGGCGCUGCCCGCACUGCCAUAAA----AUA-------CAUAGACACUUAUAGCGGUGGGGU ...(((((.........))))).........(((((.((((((.......)))))).)))))((.((((((.((((.----...-------.........)))).)))))).)). ( -37.22, z-score = -1.53, R) >droWil1.scaffold_181096 2579641 99 + 12416693 AUAUAUGCGG--GCGACGGGCAGCCCUCCAAAGGGAGUGCUUUUCCUCUCACUCUC---UACCCUUACUAUC-UAUG-----UAUGUACUAUGUAUAUAUUUUAGUCAGC----- ........((--((........)))).....((((((((..........)))))))---)...((.((((..-...(-----(((((((...))))))))..)))).)).----- ( -24.10, z-score = -0.68, R) >consensus AUCUGCCCAU__GAUGCGGGCACUUCUCCCUGCAGUUCCCUUCCCGCCCAGAAGGG___UGCCCGCACUGCCAUAAA____AUA_______CAUAGACACUUAUAGCGGUGGGGU ....((((......((((((((.....((((.....................))))...))))))))..............................((((......)))))))) (-16.26 = -17.77 + 1.50)

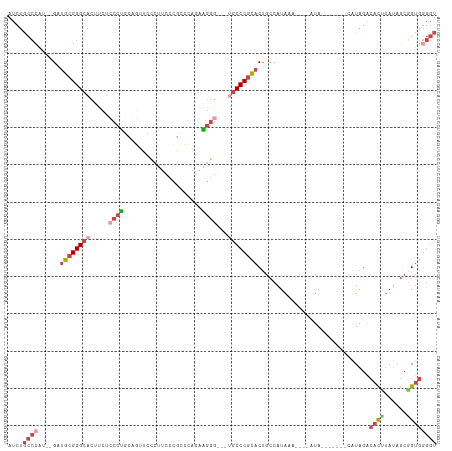
| Location | 1,125,388 – 1,125,492 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 63.83 |
| Shannon entropy | 0.66286 |
| G+C content | 0.55802 |
| Mean single sequence MFE | -39.83 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.57 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1125388 104 - 22422827 ACCCCACCGCUAUAAGUGUCUAUG-------UAU----UUUAUGGCAGUGCGGGCAGCGCCCUUCUGGGUGGGAAGGGAACUACAGGGAGAUGUGCCCGCAUCAGAUGGGCAGAU .((((((.((((((((........-------...----)))))))).))).)))....((((.((((((((((.((....))(((......))).))))).))))).)))).... ( -41.60, z-score = -1.55, R) >droEre2.scaffold_4644 1110708 106 - 2521924 ACCCCACCGCUAUCUGUGCCUAUGCGGUAUAUAUGUACUUUAUGGCUGUGCGGGC----GCCCUCUUUGCGGGAGGGAGA-GGUGGGGAGAAGUGCCCGCAUC----GGGCAGAU .(((((((.((......((((..(((((.((((.......)))))))))..))))----.((((((.....)))))))).-))))))).....((((((...)----)))))... ( -49.20, z-score = -2.27, R) >droYak2.chrX 1040171 94 - 21770863 ACCCCACAGCUUUAUGUGUCUAUGCGGUGUAUAU----UUUAUGGCAGUGCGGGC----CCCCGCUCGACGGGAGG--------GGGGUGGGGUGCCCGCAUC-----GGCAGAU .............((.((((..(((.(((.....----..))).)))((((((((----((((((((..(....).--------.)))))))).)))))))).-----)))).)) ( -44.00, z-score = -1.65, R) >droSec1.super_10 959519 104 - 3036183 ACCCCACCGCUAUAAGUGUCUAUA-------UAU----UUUAUGGCAGUGCGGGCAGCGCCCUUUUGGGCGGGAAGGGAACUGCAGGGAGAAGUGCCCGCAUCAGAUGGGCAGAU ..((((.(((((((((........-------...----)))))))).(((((((((.(((((....))))).(.((....)).).........)))))))))..).))))..... ( -41.00, z-score = -1.50, R) >droSim1.chrX_random 912481 104 - 5698898 ACCCCACCGCUAUAAGUGUCUAUG-------UAU----UUUAUGGCAGUGCGGGCAGCGCCCUUCUGGGCGGGAAGGGAACUGCAGGGAGAAGUGCCCGCAUCAGAUGGGCAGAU .((((((.((((((((........-------...----)))))))).))).)))....((((.((((((((((.((....))((........)).))))).))))).)))).... ( -41.90, z-score = -1.26, R) >droWil1.scaffold_181096 2579641 99 - 12416693 -----GCUGACUAAAAUAUAUACAUAGUACAUA-----CAUA-GAUAGUAAGGGUA---GAGAGUGAGAGGAAAAGCACUCCCUUUGGAGGGCUGCCCGUCGC--CCGCAUAUAU -----((.(((....................((-----(...-....))).(((((---(.(((((..........)))))(((....))).)))))))))))--.......... ( -21.30, z-score = 0.16, R) >consensus ACCCCACCGCUAUAAGUGUCUAUG_______UAU____UUUAUGGCAGUGCGGGCA___CCCUUCUGGGCGGGAAGGGAACUGCAGGGAGAAGUGCCCGCAUC__AUGGGCAGAU ........(((.....((((.......................))))(((((((((...((((.....(((..........))))))).....)))))))))......))).... (-17.64 = -17.57 + -0.07)

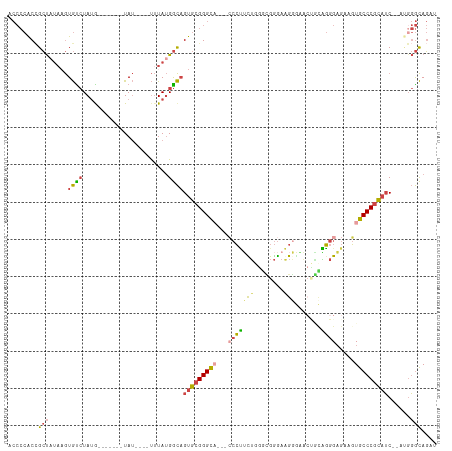
| Location | 1,125,396 – 1,125,500 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Shannon entropy | 0.37638 |
| G+C content | 0.57685 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -27.42 |
| Energy contribution | -29.86 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1125396 104 + 22422827 AUCUGAUGCGGGCACAUCUCCCUGUAGUUCCCUUCCCACCCAGAAGGGCGCUGCCCGCACUGCCAUAAA----AUA-------CAUAGACACUUAUAGCGGUGGGGUUGUGCCUU .........(((((((...(((.(((((.((((((.......)))))).)))))...((((((.((((.----...-------.........)))).))))))))).))))))). ( -39.82, z-score = -2.61, R) >droEre2.scaffold_4644 1110714 108 + 2521924 --CCGAUGCGGGCACUUCUCCCCACC-UCUCCCUCCCGCAAAGAGGGC----GCCCGCACAGCCAUAAAGUACAUAUAUACCGCAUAGGCACAGAUAGCGGUGGGGUUGUGCCUU --.......((((((....(((((((-...(((((.......))))).----((.......(((.....(((......)))......))).......)))))))))..)))))). ( -36.84, z-score = -0.84, R) >droYak2.chrX 1040176 97 + 21770863 --CCGAUGCGGGCACCCCACCC--------CCCUCCCGUCGAGCGGGG----GCCCGCACUGCCAUAAA----AUAUACACCGCAUAGACACAUAAAGCUGUGGGGUUGUGCCUU --.((.(((((((.((((.(.(--------..(....)..).).))))----))))))).)).......----...((((((.(((((.(.......)))))).)).)))).... ( -34.10, z-score = -0.51, R) >droSec1.super_10 959527 104 + 3036183 AUCUGAUGCGGGCACUUCUCCCUGCAGUUCCCUUCCCGCCCAAAAGGGCGCUGCCCGCACUGCCAUAAA----AUA-------UAUAGACACUUAUAGCGGUGGGGUUGUGCCUU .........((((((....(((.(((((.(((((.........))))).)))))...((((((.((((.----...-------.........)))).)))))))))..)))))). ( -36.42, z-score = -1.48, R) >droSim1.chrX_random 912489 104 + 5698898 AUCUGAUGCGGGCACUUCUCCCUGCAGUUCCCUUCCCGCCCAGAAGGGCGCUGCCCGCACUGCCAUAAA----AUA-------CAUAGACACUUAUAGCGGUGGGGUUGUGCCCU .........((((((....(((.(((((.((((((.......)))))).)))))...((((((.((((.----...-------.........)))).)))))))))..)))))). ( -42.92, z-score = -3.01, R) >consensus AUCUGAUGCGGGCACUUCUCCCUGCAGUUCCCUUCCCGCCCAGAAGGGCGCUGCCCGCACUGCCAUAAA____AUA_______CAUAGACACUUAUAGCGGUGGGGUUGUGCCUU .........((((((........(((((.((((((.......)))))).)))))((.((((((.((((........................)))).)))))).))..)))))). (-27.42 = -29.86 + 2.44)


| Location | 1,125,396 – 1,125,500 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Shannon entropy | 0.37638 |
| G+C content | 0.57685 |
| Mean single sequence MFE | -43.67 |
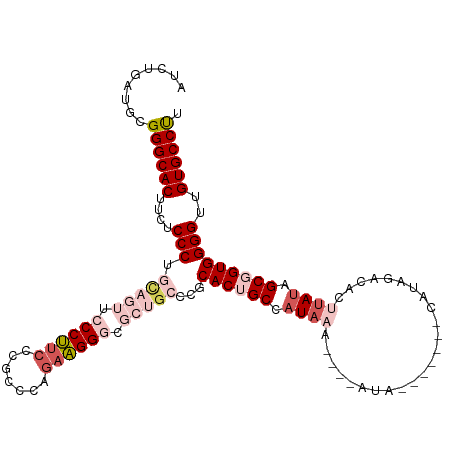
| Consensus MFE | -31.15 |
| Energy contribution | -32.18 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1125396 104 - 22422827 AAGGCACAACCCCACCGCUAUAAGUGUCUAUG-------UAU----UUUAUGGCAGUGCGGGCAGCGCCCUUCUGGGUGGGAAGGGAACUACAGGGAGAUGUGCCCGCAUCAGAU ..((((((.((((.((((....(.((.(((((-------...----..))))))).)))))(.((..(((((((.....)))))))..)).).))).).)))))).......... ( -37.60, z-score = -0.81, R) >droEre2.scaffold_4644 1110714 108 - 2521924 AAGGCACAACCCCACCGCUAUCUGUGCCUAUGCGGUAUAUAUGUACUUUAUGGCUGUGCGGGC----GCCCUCUUUGCGGGAGGGAGA-GGUGGGGAGAAGUGCCCGCAUCGG-- ..(((((..(((((((.((......((((..(((((.((((.......)))))))))..))))----.((((((.....)))))))).-)))))))....)))))........-- ( -47.30, z-score = -1.82, R) >droYak2.chrX 1040176 97 - 21770863 AAGGCACAACCCCACAGCUUUAUGUGUCUAUGCGGUGUAUAU----UUUAUGGCAGUGCGGGC----CCCCGCUCGACGGGAGGG--------GGGUGGGGUGCCCGCAUCGG-- .(((((((..............))))))).(((.(((.....----..))).)))((((((((----((((((((..(....)..--------)))))))).))))))))...-- ( -46.24, z-score = -2.11, R) >droSec1.super_10 959527 104 - 3036183 AAGGCACAACCCCACCGCUAUAAGUGUCUAUA-------UAU----UUUAUGGCAGUGCGGGCAGCGCCCUUUUGGGCGGGAAGGGAACUGCAGGGAGAAGUGCCCGCAUCAGAU ..(((((..((((.((((....(.((.(((((-------...----..))))))).)))))((((..(((((((.....)))))))..)))).))).)..))))).......... ( -40.30, z-score = -1.68, R) >droSim1.chrX_random 912489 104 - 5698898 AGGGCACAACCCCACCGCUAUAAGUGUCUAUG-------UAU----UUUAUGGCAGUGCGGGCAGCGCCCUUCUGGGCGGGAAGGGAACUGCAGGGAGAAGUGCCCGCAUCAGAU .((((((..((((.((((....(.((.(((((-------...----..))))))).)))))((((..(((((((.....)))))))..)))).))).)..))))))......... ( -46.90, z-score = -2.82, R) >consensus AAGGCACAACCCCACCGCUAUAAGUGUCUAUG_______UAU____UUUAUGGCAGUGCGGGCAGCGCCCUUCUGGGCGGGAAGGGAACUGCAGGGAGAAGUGCCCGCAUCAGAU ..(((((..((((.((((......((((.......................))))..))))((((..(((((((.....)))))))..)))).))).)..))))).......... (-31.15 = -32.18 + 1.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:28 2011