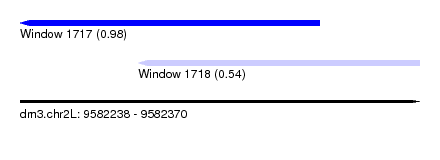
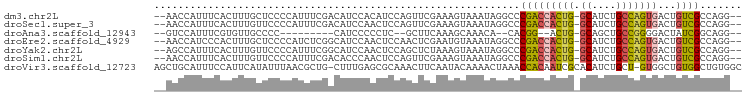
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,582,238 – 9,582,370 |
| Length | 132 |
| Max. P | 0.984566 |

| Location | 9,582,238 – 9,582,337 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 68.31 |
| Shannon entropy | 0.61736 |
| G+C content | 0.54542 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -13.15 |
| Energy contribution | -14.56 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9582238 99 - 23011544 ACAUCCAGUUCGAAAGUAAAUAGGCCCGACCACUGGCAUCUGCCAGUGACUGUCGC--CAGGUGUACCCAUCAGAACCCAUGCCAUACAGCACAUCCCCAU .......((((...........(((..((((((((((....)))))))...)))))--).((((....)))).))))...(((......)))......... ( -26.10, z-score = -2.21, R) >droSec1.super_3 5029377 99 - 7220098 AACUCCAGUUCGAAAGUAAAUAGGCCCGACCACUGGCAUCUGCCAGUGACUGUCGC--CAGGUGUACCCAUCAGAACCCAUGCCAUACAGCAGAUCCCCAU .......((((...........(((..((((((((((....)))))))...)))))--).((((....)))).))))...(((......)))......... ( -26.40, z-score = -2.19, R) >droAna3.scaffold_12943 998244 80 - 5039921 --CCCUCGCUUCAAAGCAAACA--CACGG--ACUGGCAGCUGCCGGGGACUAUCGG--CAGGUGUACCCAUCGCAGCUCCUGCCAUCC------------- --.....(((....))).....--...((--(.((((((((((..(((((.(((..--..))))).)))...)))))...))))))))------------- ( -27.70, z-score = -1.30, R) >droEre2.scaffold_4929 10187967 88 - 26641161 AACUCCAACUCGAAUGUAAAUAGGCCCGACCACUGGCAUCUGCCAGUGACUGUCGC--CAGGUGUACCCAUCAGAACCGAUGCCAUACAG----------- ..............((((..((.((((((((((((((....)))))))...)))).--..))).))..((((......))))...)))).----------- ( -24.40, z-score = -2.10, R) >droYak2.chr2L 12265821 82 - 22324452 AACUCCAGCUCUAAAGUAAAUAGGCCCGACCACUGGCAUCUGCCAGUGACUGUCGC--CAGGUGUACCCAUCAGAUCCCCAUCU----------------- .((.((.(((....))).....(((..((((((((((....)))))))...)))))--).)).))...................----------------- ( -21.90, z-score = -1.93, R) >droSim1.chr2L 9356079 99 - 22036055 AACUCCAGUUCGAAAGUAAAUAGGCCCGACCACUGGCAUCUGCCAGUGACUGUCGC--CAGGUGUACCCAUCAGAACCCAUGCCAUAUAGCAGAUCCCCAU .......((((...........(((..((((((((((....)))))))...)))))--).((((....)))).))))...(((......)))......... ( -26.40, z-score = -2.32, R) >droVir3.scaffold_12723 4060804 95 - 5802038 AACUUCAAUACAAAACUAAAC---CACAAUCGC--ACAUCUGCU-GUGGCUGUGGCUGUGGCUGCUCUGACUACGCCUGCGACUGUGCUCCAGGUCCUGGU .....................---((((.((((--(.....((.-(((((.(..((....))..)...).)))))).))))).))))..((((...)))). ( -23.10, z-score = 0.09, R) >consensus AACUCCAGUUCGAAAGUAAAUAGGCCCGACCACUGGCAUCUGCCAGUGACUGUCGC__CAGGUGUACCCAUCAGAACCCAUGCCAUACAGCA__UCC___U ......................(((.(((((((((((....)))))))...)))).....((((....)))).........)))................. (-13.15 = -14.56 + 1.41)
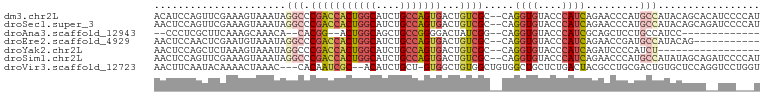
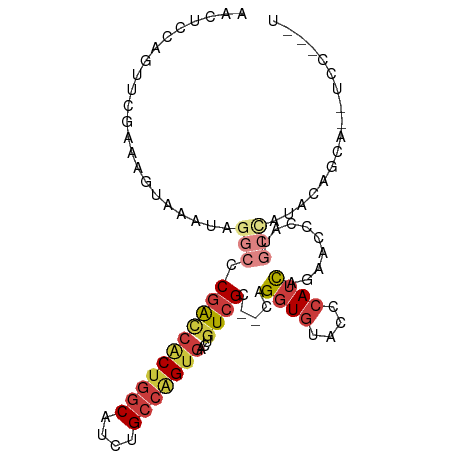
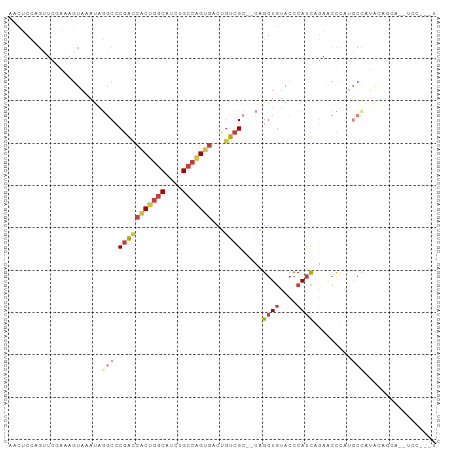
| Location | 9,582,277 – 9,582,370 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 71.54 |
| Shannon entropy | 0.56864 |
| G+C content | 0.52671 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -8.88 |
| Energy contribution | -9.03 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9582277 93 - 23011544 --AACCAUUUCACUUUGCUCCCCAUUUCGACAUCCACAUCCAGUUCGAAAGUAAAUAGGCCCGACCACUG-GCAUCUGCCAGUGACUGUCGCCAGG-- --..((.......((((((......(((((..............)))))))))))..(((..((((((((-((....)))))))...)))))).))-- ( -23.34, z-score = -2.49, R) >droSec1.super_3 5029416 93 - 7220098 --AACCAUUUCACUUUGUUCCCCAUUUCGACAUCCAACUCCAGUUCGAAAGUAAAUAGGCCCGACCACUG-GCAUCUGCCAGUGACUGUCGCCAGG-- --..((..................((((((..............)))))).......(((..((((((((-((....)))))))...)))))).))-- ( -22.64, z-score = -2.25, R) >droAna3.scaffold_12943 998270 78 - 5039921 --GUCCAUUUCGUGUUGCCCC---------CAUCCCCCUC--GCUUCAAAGCAAACA--CACGG--ACUG-GCAGCUGCCGGGGACUAUCGGCAGG-- --(.(((.((((((((((...---------..........--........)))...)--)))))--).))-))..(((((((......))))))).-- ( -23.50, z-score = -0.37, R) >droEre2.scaffold_4929 10187995 93 - 26641161 --AACCAUCCCACUUUGCUCCCCAUCUCGGCAUCCAACUCCAACUCGAAUGUAAAUAGGCCCGACCACUG-GCAUCUGCCAGUGACUGUCGCCAGG-- --..((.........((((.........)))).........................(((..((((((((-((....)))))))...)))))).))-- ( -21.30, z-score = -1.34, R) >droYak2.chr2L 12265843 93 - 22324452 --AGCCAUUUCACUUUGUUCCCCAUUUCGGCAUCCAACUCCAGCUCUAAAGUAAAUAGGCCCGACCACUG-GCAUCUGCCAGUGACUGUCGCCAGG-- --.(((((((.((((((...........((...))...........)))))))))).))).(((((((((-((....)))))))...)))).....-- ( -22.95, z-score = -1.56, R) >droSim1.chr2L 9356118 93 - 22036055 --AACCAUUUCACUUUGUUCCCCAUUUCGACACCCAACUCCAGUUCGAAAGUAAAUAGGCCCGACCACUG-GCAUCUGCCAGUGACUGUCGCCAGG-- --..((..................((((((.((.........)))))))).......(((..((((((((-((....)))))))...)))))).))-- ( -23.20, z-score = -2.43, R) >droVir3.scaffold_12723 4060843 96 - 5802038 AGCUGCAUUUCCAUUCAUAUUUAACGCUG-CUUUGAGCGCAAACUUCAAUACAAAACUAAACCACAAUCGCACAUCUGCU-GUGGCUGUGGCUGUGGC .((..((.................((((.-.....))))......................(((((..(((((....).)-)))..))))).))..)) ( -21.80, z-score = -0.12, R) >consensus __AACCAUUUCACUUUGUUCCCCAUUUCGACAUCCAACUCCAGCUCGAAAGUAAAUAGGCCCGACCACUG_GCAUCUGCCAGUGACUGUCGCCAGG__ .............................................................(((((((((.((....)))))))...))))....... ( -8.88 = -9.03 + 0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:33 2011