
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 844,341 – 844,442 |
| Length | 101 |
| Max. P | 0.706431 |

| Location | 844,341 – 844,442 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Shannon entropy | 0.33899 |
| G+C content | 0.47398 |
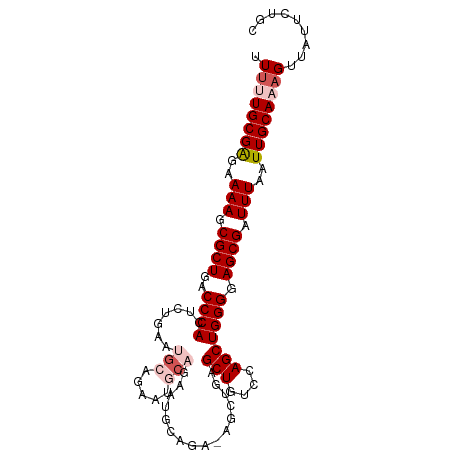
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -19.89 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
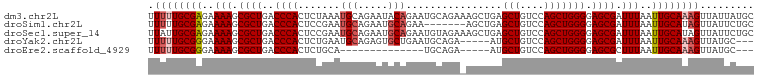
>dm3.chr2L 844341 101 + 23011544 UUUUUGCGAGAAAAGCGCUGACCCACUCUAAAUGCAGAAUACAGAAUGCAGAAAGCUGAGCUGUCCAGCUGGGGAGCGAUUUAAUUGCAAAGUUAUUAUGC .((((((((..(((.((((..(((........((((..........))))...(((((.......)))))))).)))).)))..))))))))......... ( -28.70, z-score = -1.58, R) >droSim1.chr2L 843221 94 + 22036055 UUUUUGCGAGAAAAGCGCUGACCCACUCCGAAUGCAGAAUGCAGAA-------AGCUGAGCUGUCCAGCUGGGGAGCGAUUUAAUUGCAUAGUUAUUCUGC ....(((((..(((.((((..(((........(((.....)))...-------(((((.......)))))))).)))).)))..)))))............ ( -25.80, z-score = -0.35, R) >droSec1.super_14 814983 101 + 2068291 UUAUUGCGAGAAAAGCGCUGACCCACUCCGAAUGCAGAAUGCAGAAUGUAGAAAGCUGAGCUGUCCAGCUGGGGAGCGAUUUAAUUGCAUAGUUAUUCUGC ....(((((..(((.((((..(((.((.((..(((.....)))...)).))..(((((.......)))))))).)))).)))..)))))............ ( -26.60, z-score = -0.45, R) >droYak2.chr2L 827948 93 + 22324452 UUUUUGCGGGAAAAGCGCUGACCCACUCUGAAUGCAGAGUGCUGAAUGCAGA-----AUGCUGUCCAGCUGGGGAGCGAUUUAAUUGCAAAGUUAUGC--- .((((((((..(((.((((..(((((((((....))))))((((...((((.-----...)))).)))).))).)))).)))..))))))))......--- ( -34.20, z-score = -2.83, R) >droEre2.scaffold_4929 901050 79 + 26641161 UUUUUGCGGGAAAAGCGCUGACCCACUCUGCA--------------UGCAGA-----AUGCUGUCCAGCUGGGGAGCGCUUUAAUUGCAAAGUUAUGC--- .((((((((..((((((((..((((.((((..--------------..))))-----..(((....))))))).))))))))..))))))))......--- ( -32.50, z-score = -3.05, R) >consensus UUUUUGCGAGAAAAGCGCUGACCCACUCUGAAUGCAGAAUGCAGAAUGCAGA_AGCUGAGCUGUCCAGCUGGGGAGCGAUUUAAUUGCAAAGUUAUUCUGC .((((((((..(((.((((..((((.......(((.....)))................(((....))))))).)))).)))..))))))))......... (-19.89 = -20.80 + 0.91)
| Location | 844,341 – 844,442 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Shannon entropy | 0.33899 |
| G+C content | 0.47398 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -17.76 |
| Energy contribution | -19.24 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.706431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 844341 101 - 23011544 GCAUAAUAACUUUGCAAUUAAAUCGCUCCCCAGCUGGACAGCUCAGCUUUCUGCAUUCUGUAUUCUGCAUUUAGAGUGGGUCAGCGCUUUUCUCGCAAAAA ..........(((((....(((.((((..((((((((.....)))))).....(((((((...........)))))))))..)))).)))....))))).. ( -25.10, z-score = -1.05, R) >droSim1.chr2L 843221 94 - 22036055 GCAGAAUAACUAUGCAAUUAAAUCGCUCCCCAGCUGGACAGCUCAGCU-------UUCUGCAUUCUGCAUUCGGAGUGGGUCAGCGCUUUUCUCGCAAAAA ............(((....(((.((((..((((((((.....))))))-------.....(((((((....)))))))))..)))).)))....))).... ( -24.30, z-score = -0.43, R) >droSec1.super_14 814983 101 - 2068291 GCAGAAUAACUAUGCAAUUAAAUCGCUCCCCAGCUGGACAGCUCAGCUUUCUACAUUCUGCAUUCUGCAUUCGGAGUGGGUCAGCGCUUUUCUCGCAAUAA ............(((....(((.((((..((((((((.....))))))............(((((((....)))))))))..)))).)))....))).... ( -24.30, z-score = -0.45, R) >droYak2.chr2L 827948 93 - 22324452 ---GCAUAACUUUGCAAUUAAAUCGCUCCCCAGCUGGACAGCAU-----UCUGCAUUCAGCACUCUGCAUUCAGAGUGGGUCAGCGCUUUUCCCGCAAAAA ---.......(((((....(((.((((.(((.((((((..((..-----...)).))))))((((((....)))))))))..)))).)))....))))).. ( -30.30, z-score = -3.20, R) >droEre2.scaffold_4929 901050 79 - 26641161 ---GCAUAACUUUGCAAUUAAAGCGCUCCCCAGCUGGACAGCAU-----UCUGCA--------------UGCAGAGUGGGUCAGCGCUUUUCCCGCAAAAA ---.......(((((....((((((((.(((.(((....)))((-----((((..--------------..)))))))))..))))))))....))))).. ( -30.50, z-score = -3.33, R) >consensus GCAGAAUAACUUUGCAAUUAAAUCGCUCCCCAGCUGGACAGCUCAGCU_UCUGCAUUCUGCAUUCUGCAUUCAGAGUGGGUCAGCGCUUUUCUCGCAAAAA ..........(((((....(((.((((..((.(((....)))..................(((((((....)))))))))..)))).)))....))))).. (-17.76 = -19.24 + 1.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:10 2011