
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,116,196 – 1,116,295 |
| Length | 99 |
| Max. P | 0.684117 |

| Location | 1,116,196 – 1,116,295 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.78 |
| Shannon entropy | 0.27909 |
| G+C content | 0.44736 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.54 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
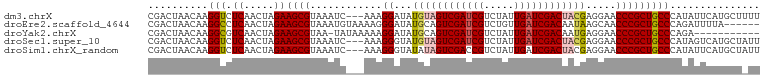
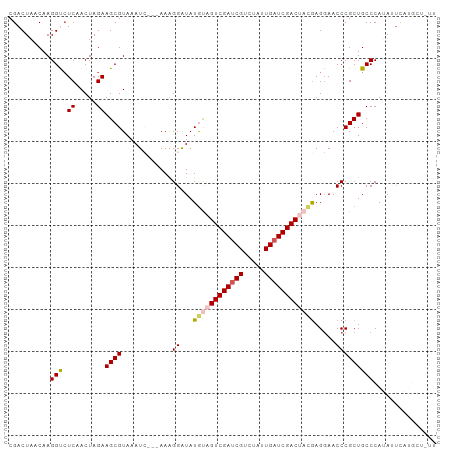
>dm3.chrX 1116196 99 + 22422827 CGACUAACAAGGUCUCAACUAGAAGCGUAAAUC---AAAGGAUAUGUAGUCGAUCGUCUAUUGAUCGACUACGAGGAACCCGCUGCCCAUAUUCAUGCUUUU .((((.....))))......((((((((.....---...(((((((((((((((((.....)))))))))))..((..........)))))))))))))))) ( -28.20, z-score = -2.74, R) >droEre2.scaffold_4644 1101480 96 + 2521924 CGACUAACAAGGCCUCAACUAGAAGCGUAAAUGUAAAAGGGAUAUGCAGUCGAUCGUCUGUUGAUCGACAAUAAGCAACCCGCUGCCCAGAUUUUA------ ...........((.((.....)).))............(((.......((((((((.....))))))))....(((.....))).)))........------ ( -21.10, z-score = -0.62, R) >droYak2.chrX 1030915 90 + 21770863 CGACUAACAAGGCGUCAACUAGAAGCGUAA-UAUAAAAAGGAUAUGCAGUCGAUCGUCUAUUGAUCGACAAUGAGGAACCCGCUGCCCAGA----------- ..........((((.(........(((((.-...........))))).((((((((.....))))))))............).))))....----------- ( -20.10, z-score = -0.84, R) >droSec1.super_10 950402 99 + 3036183 CGACUAACAAGGUCUCAACUAGAAGCGUAAAUC---AAAGGGUAUGUAGUCGAUCGUCUAUUGAUCGACUACGAGGAACCCGCUGCCCAUAGUCAUGCUAUU .(((((.....(..((.....))..).......---...(((((((((((((((((.....)))))))))))).((...))..))))).)))))........ ( -31.70, z-score = -3.25, R) >droSim1.chrX_random 899771 99 + 5698898 CGACUAACAAGGUCUCAACUAGAAGCGUAAAUC---AAAGGGUAUAUAGUCGACCGUCUAUUGAUCGACUACGAGGAACCCGCUGCCCAUAUUCAUGCUAUU .((((.....)))).........(((((.(((.---...(((((..(((((((.((.....)).)))))))((.(....))).)))))..))).)))))... ( -23.20, z-score = -1.13, R) >consensus CGACUAACAAGGUCUCAACUAGAAGCGUAAAUC___AAAGGAUAUGUAGUCGAUCGUCUAUUGAUCGACUACGAGGAACCCGCUGCCCAUAUUCAUGCU_UU ..........(((.((.....))((((.................((((((((((((.....)))))))))))).(....))))))))............... (-16.90 = -17.54 + 0.64)

| Location | 1,116,196 – 1,116,295 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.78 |
| Shannon entropy | 0.27909 |
| G+C content | 0.44736 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
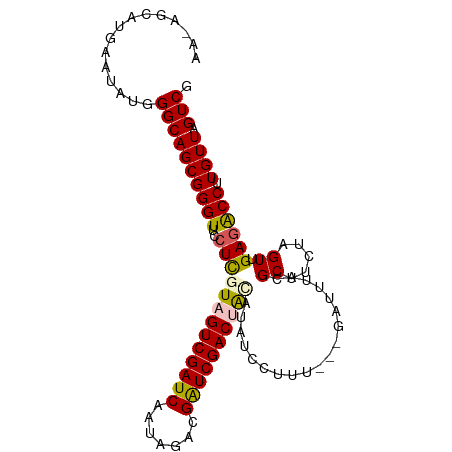
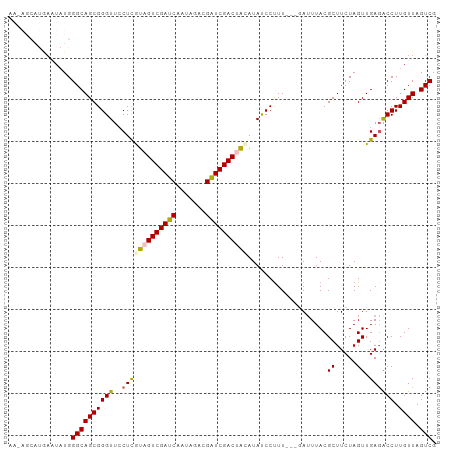
>dm3.chrX 1116196 99 - 22422827 AAAAGCAUGAAUAUGGGCAGCGGGUUCCUCGUAGUCGAUCAAUAGACGAUCGACUACAUAUCCUUU---GAUUUACGCUUCUAGUUGAGACCUUGUUAGUCG ..((((.(((((..(((....(((...)))((((((((((.......))))))))))....)))..---.))))).))))........(((.......))). ( -26.50, z-score = -1.36, R) >droEre2.scaffold_4644 1101480 96 - 2521924 ------UAAAAUCUGGGCAGCGGGUUGCUUAUUGUCGAUCAACAGACGAUCGACUGCAUAUCCCUUUUACAUUUACGCUUCUAGUUGAGGCCUUGUUAGUCG ------.........(((((((((.(((.....(((((((.......))))))).)))...)))............(((((.....)))))...))).))). ( -27.70, z-score = -2.15, R) >droYak2.chrX 1030915 90 - 21770863 -----------UCUGGGCAGCGGGUUCCUCAUUGUCGAUCAAUAGACGAUCGACUGCAUAUCCUUUUUAUA-UUACGCUUCUAGUUGACGCCUUGUUAGUCG -----------.(((((.(((((....).....(((((((.......))))))).................-...)))))))))..(((.........))). ( -20.30, z-score = -0.35, R) >droSec1.super_10 950402 99 - 3036183 AAUAGCAUGACUAUGGGCAGCGGGUUCCUCGUAGUCGAUCAAUAGACGAUCGACUACAUACCCUUU---GAUUUACGCUUCUAGUUGAGACCUUGUUAGUCG ........(((((.((.((((((((.....((((((((((.......))))))))))..))))...---((........))..))))...))....))))). ( -32.20, z-score = -2.99, R) >droSim1.chrX_random 899771 99 - 5698898 AAUAGCAUGAAUAUGGGCAGCGGGUUCCUCGUAGUCGAUCAAUAGACGGUCGACUAUAUACCCUUU---GAUUUACGCUUCUAGUUGAGACCUUGUUAGUCG .........((((.((.((((((((.....((((((((((.......))))))))))..))))...---((........))..))))...)).))))..... ( -25.80, z-score = -0.81, R) >consensus AA_AGCAUGAAUAUGGGCAGCGGGUUCCUCGUAGUCGAUCAAUAGACGAUCGACUACAUAUCCUUU___GAUUUACGCUUCUAGUUGAGACCUUGUUAGUCG ...............((((((((((..(((((((((((((.......))))))))))...................((.....)).)))))).)))).))). (-20.38 = -20.50 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:22 2011