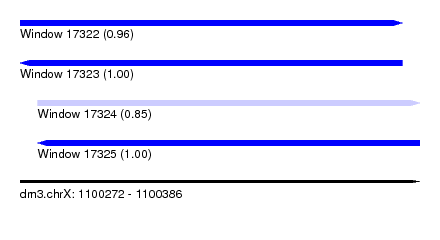
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,100,272 – 1,100,386 |
| Length | 114 |
| Max. P | 0.999673 |

| Location | 1,100,272 – 1,100,381 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.33160 |
| G+C content | 0.36572 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -9.92 |
| Energy contribution | -11.44 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1100272 109 + 22422827 GGGAAAACGUCUACACAGAAAAAAA----CAUUACGAUCUCUUGUGGCUACUGAACGCAAGAAAUUCAUGUUGAUUAUGACCAUGUUAUAGCAAUUUUUU-CUGUGCAGCACGG .......(((((.(((((((((((.----......((..(((((((.........)))))))...))...(((.(((((((...))))))))))))))))-))))).)).))). ( -30.00, z-score = -3.32, R) >droSim1.chrX 879374 109 + 17042790 GGAAAACCGUCUACACAGAAAAAAA----CUUUACGAUCUCUUGCGGCUACUUUAUGCAAGAAAUUCAUGUUGAUUAUCACCAUGUUAUGUCAAUUUUUU-CUGUGCAGCACGG ......((((((.(((((((((((.----......(((.(((((((.........))))))).)))((((.((.....)).)))).........))))))-))))).)).)))) ( -30.30, z-score = -4.13, R) >droSec1.super_10 931837 109 + 3036183 GGAAAACCGUCUACACAGAAAAAAA----CUUUACGAUCUCUUGUGGCUAUUUAAUGCAAGAAAUUCAUGUUGAUUAUCACCAUGUUAUGUCAAUUUUUU-CUGUGCAGCACGG ......((((((.(((((((((((.----......(((.(((((((.........))))))).)))((((.((.....)).)))).........))))))-))))).)).)))) ( -27.70, z-score = -3.10, R) >droYak2.chrX 1014910 109 + 21770863 GGAAAACCGCCUACACAGAAAAAAA----CAUAACCAUCUCUUUUGGUUACUAAAUGGUAGAGAUUCAUGUUGAUUAUCACCACAUUAGAGCAUUUUUUUUCUGUGCAGCCAG- ((....))(.((.((((((((((((----..((((((.......))))))((((.((((.((...((.....))...))))))..)))).....)))))))))))).)).)..- ( -27.90, z-score = -3.63, R) >droEre2.scaffold_4644 1085271 111 + 2521924 GCAAAACAGCCUGCACAGAAAAAAAAAAACAUUAACAUGAAC-AUGCCUACUAAAUGAAAGAAAUUCAUGUUGAAUAUUACCAUAUUAUAGCAGUUUUUU-CCGUGCAGGCGG- ........((((((((.(((((((........((((((((((-((.........))).......)))))))))(((((....))))).......))))))-).))))))))..- ( -28.71, z-score = -4.46, R) >consensus GGAAAACCGUCUACACAGAAAAAAA____CAUUACGAUCUCUUGUGGCUACUAAAUGCAAGAAAUUCAUGUUGAUUAUCACCAUGUUAUAGCAAUUUUUU_CUGUGCAGCACGG ..........((.(((((((((((...............(((((((.........)))))))....((((.((.....)).)))).........)))))).))))).))..... ( -9.92 = -11.44 + 1.52)

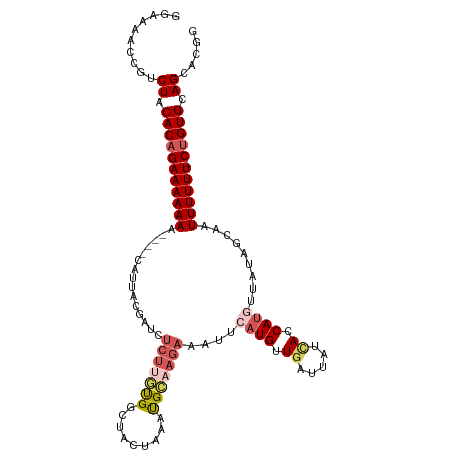
| Location | 1,100,272 – 1,100,381 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.33160 |
| G+C content | 0.36572 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -18.80 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.23 |
| Mean z-score | -4.91 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1100272 109 - 22422827 CCGUGCUGCACAG-AAAAAAUUGCUAUAACAUGGUCAUAAUCAACAUGAAUUUCUUGCGUUCAGUAGCCACAAGAGAUCGUAAUG----UUUUUUUCUGUGUAGACGUUUUCCC .(((.((((((((-((((((((((.....((((...........)))).((((((((.((......))..)))))))).))))).----..))))))))))))))))....... ( -31.00, z-score = -3.73, R) >droSim1.chrX 879374 109 - 17042790 CCGUGCUGCACAG-AAAAAAUUGACAUAACAUGGUGAUAAUCAACAUGAAUUUCUUGCAUAAAGUAGCCGCAAGAGAUCGUAAAG----UUUUUUUCUGUGUAGACGGUUUUCC ((((.((((((((-((((((...((....((((.((.....)).)))).(((((((((...........))))))))).))....----.))))))))))))))))))...... ( -37.90, z-score = -5.61, R) >droSec1.super_10 931837 109 - 3036183 CCGUGCUGCACAG-AAAAAAUUGACAUAACAUGGUGAUAAUCAACAUGAAUUUCUUGCAUUAAAUAGCCACAAGAGAUCGUAAAG----UUUUUUUCUGUGUAGACGGUUUUCC ((((.((((((((-((((((...((....((((.((.....)).)))).((((((((.............)))))))).))....----.))))))))))))))))))...... ( -33.92, z-score = -4.66, R) >droYak2.chrX 1014910 109 - 21770863 -CUGGCUGCACAGAAAAAAAAUGCUCUAAUGUGGUGAUAAUCAACAUGAAUCUCUACCAUUUAGUAACCAAAAGAGAUGGUUAUG----UUUUUUUCUGUGUAGGCGGUUUUCC -(((.(((((((((((((((.....((((.(((((((.(.((.....)).).)).)))))))))((((((.......))))))..----))))))))))))))).)))...... ( -35.20, z-score = -4.65, R) >droEre2.scaffold_4644 1085271 111 - 2521924 -CCGCCUGCACGG-AAAAAACUGCUAUAAUAUGGUAAUAUUCAACAUGAAUUUCUUUCAUUUAGUAGGCAU-GUUCAUGUUAAUGUUUUUUUUUUUCUGUGCAGGCUGUUUUGC -..((((((((((-((((((.((((((...))))))(((((.(((((((((..(((.(.....).)))...-))))))))))))))....))))))))))))))))........ ( -38.10, z-score = -5.90, R) >consensus CCGUGCUGCACAG_AAAAAAUUGCUAUAACAUGGUGAUAAUCAACAUGAAUUUCUUGCAUUAAGUAGCCACAAGAGAUCGUAAUG____UUUUUUUCUGUGUAGACGGUUUUCC .....((((((((.((((((.........((((...........)))).((((((((.............))))))))...........)))))).)))))))).......... (-18.80 = -19.20 + 0.40)

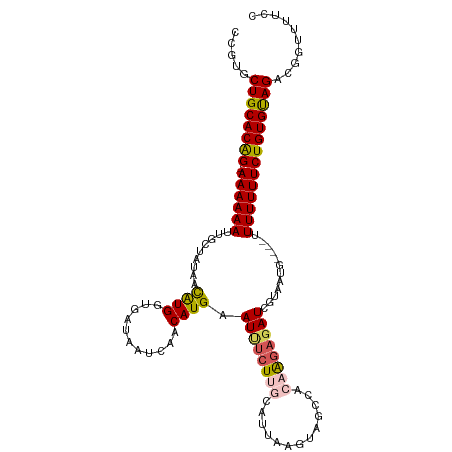
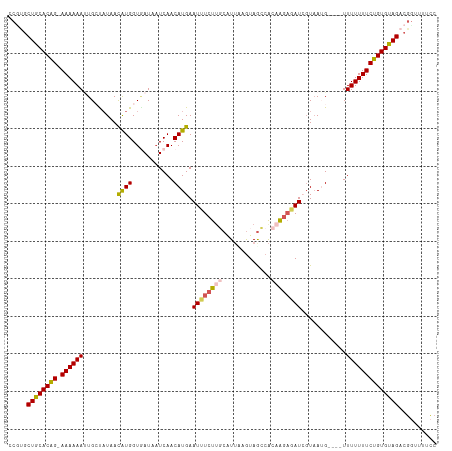
| Location | 1,100,277 – 1,100,386 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.51 |
| Shannon entropy | 0.30472 |
| G+C content | 0.37302 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -10.50 |
| Energy contribution | -13.02 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1100277 109 + 22422827 AACGUCUACACAGAAAAAAA----CAUUACGAUCUCUUGUGGCUACUGAACGCAAGAAAUUCAUGUUGAUUAUGACCAUGUUAUAGCAAUUUUUU-CUGUGCAGCACGGCAGCU ..(((((.(((((((((((.----......((..(((((((.........)))))))...))...(((.(((((((...))))))))))))))))-))))).)).)))...... ( -30.00, z-score = -2.80, R) >droSim1.chrX 879379 109 + 17042790 ACCGUCUACACAGAAAAAAA----CUUUACGAUCUCUUGCGGCUACUUUAUGCAAGAAAUUCAUGUUGAUUAUCACCAUGUUAUGUCAAUUUUUU-CUGUGCAGCACGGCAGCU .((((((.(((((((((((.----......(((.(((((((.........))))))).)))((((.((.....)).)))).........))))))-))))).)).))))..... ( -30.70, z-score = -3.80, R) >droSec1.super_10 931842 109 + 3036183 ACCGUCUACACAGAAAAAAA----CUUUACGAUCUCUUGUGGCUAUUUAAUGCAAGAAAUUCAUGUUGAUUAUCACCAUGUUAUGUCAAUUUUUU-CUGUGCAGCACGGCAGCU .((((((.(((((((((((.----......(((.(((((((.........))))))).)))((((.((.....)).)))).........))))))-))))).)).))))..... ( -28.10, z-score = -2.84, R) >droYak2.chrX 1014915 109 + 21770863 ACCGCCUACACAGAAAAAAA----CAUAACCAUCUCUUUUGGUUACUAAAUGGUAGAGAUUCAUGUUGAUUAUCACCACAUUAGAGCAUUUUUUUUCUGUGCAGC-CAGCAGCU ...((((.((((((((((((----..((((((.......))))))((((.((((.((...((.....))...))))))..)))).....)))))))))))).)).-..)).... ( -28.00, z-score = -3.46, R) >droEre2.scaffold_4644 1085276 111 + 2521924 ACAGCCUGCACAGAAAAAAAAAAACAUUAACAUGAAC-AUGCCUACUAAAUGAAAGAAAUUCAUGUUGAAUAUUACCAUAUUAUAGCAGUUUUUU-CCGUGCAGG-CGGCAGCU ...((((((((.(((((((........((((((((((-((.........))).......)))))))))(((((....))))).......))))))-).)))))))-)....... ( -28.71, z-score = -3.66, R) >consensus ACCGUCUACACAGAAAAAAA____CAUUACGAUCUCUUGUGGCUACUAAAUGCAAGAAAUUCAUGUUGAUUAUCACCAUGUUAUAGCAAUUUUUU_CUGUGCAGCACGGCAGCU .((((((.(((((((((((...............(((((((.........)))))))....((((.((.....)).)))).........)))))).))))).)).))))..... (-10.50 = -13.02 + 2.52)

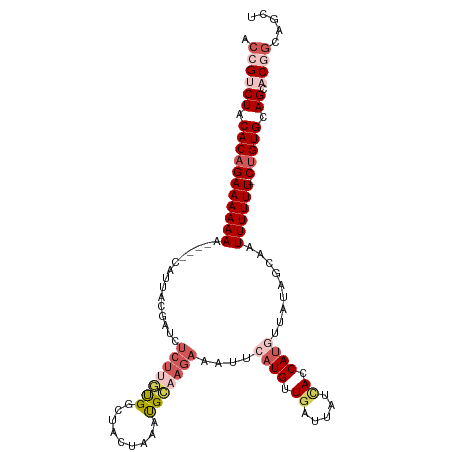
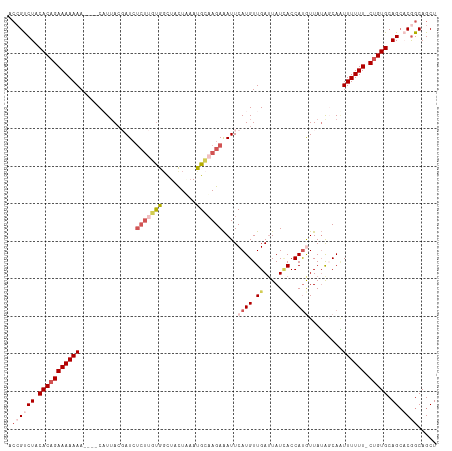
| Location | 1,100,277 – 1,100,386 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.51 |
| Shannon entropy | 0.30472 |
| G+C content | 0.37302 |
| Mean single sequence MFE | -36.24 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.92 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1100277 109 - 22422827 AGCUGCCGUGCUGCACAG-AAAAAAUUGCUAUAACAUGGUCAUAAUCAACAUGAAUUUCUUGCGUUCAGUAGCCACAAGAGAUCGUAAUG----UUUUUUUCUGUGUAGACGUU ......(((.((((((((-((((((((((.....((((...........)))).((((((((.((......))..)))))))).))))).----..)))))))))))))))).. ( -31.00, z-score = -2.51, R) >droSim1.chrX 879379 109 - 17042790 AGCUGCCGUGCUGCACAG-AAAAAAUUGACAUAACAUGGUGAUAAUCAACAUGAAUUUCUUGCAUAAAGUAGCCGCAAGAGAUCGUAAAG----UUUUUUUCUGUGUAGACGGU ....(((((.((((((((-((((((...((....((((.((.....)).)))).(((((((((...........))))))))).))....----.))))))))))))))))))) ( -39.50, z-score = -5.29, R) >droSec1.super_10 931842 109 - 3036183 AGCUGCCGUGCUGCACAG-AAAAAAUUGACAUAACAUGGUGAUAAUCAACAUGAAUUUCUUGCAUUAAAUAGCCACAAGAGAUCGUAAAG----UUUUUUUCUGUGUAGACGGU ....(((((.((((((((-((((((...((....((((.((.....)).)))).((((((((.............)))))))).))....----.))))))))))))))))))) ( -35.52, z-score = -4.46, R) >droYak2.chrX 1014915 109 - 21770863 AGCUGCUG-GCUGCACAGAAAAAAAAUGCUCUAAUGUGGUGAUAAUCAACAUGAAUCUCUACCAUUUAGUAACCAAAAGAGAUGGUUAUG----UUUUUUUCUGUGUAGGCGGU ....((((-.(((((((((((((((.....((((.(((((((.(.((.....)).).)).)))))))))((((((.......))))))..----))))))))))))))).)))) ( -36.80, z-score = -4.50, R) >droEre2.scaffold_4644 1085276 111 - 2521924 AGCUGCCG-CCUGCACGG-AAAAAACUGCUAUAAUAUGGUAAUAUUCAACAUGAAUUUCUUUCAUUUAGUAGGCAU-GUUCAUGUUAAUGUUUUUUUUUUUCUGUGCAGGCUGU ....((.(-(((((((((-((((((.((((((...))))))(((((.(((((((((..(((.(.....).)))...-))))))))))))))....)))))))))))))))).)) ( -38.40, z-score = -5.34, R) >consensus AGCUGCCGUGCUGCACAG_AAAAAAUUGCUAUAACAUGGUGAUAAUCAACAUGAAUUUCUUGCAUUAAGUAGCCACAAGAGAUCGUAAUG____UUUUUUUCUGUGUAGACGGU ....(((((.((((((((.((((((.........((((...........)))).((((((((.............))))))))...........)))))).))))))))))))) (-20.84 = -21.92 + 1.08)

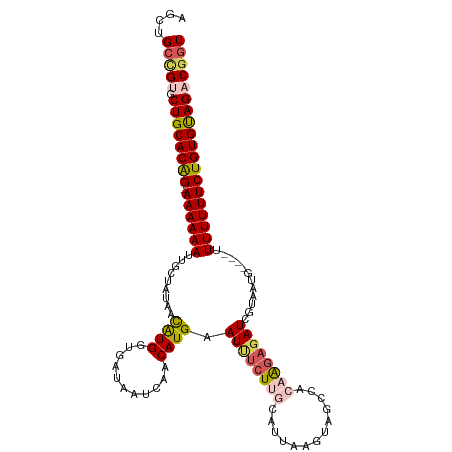
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:19 2011