| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,072,845 – 1,072,919 |
| Length | 74 |
| Max. P | 0.991502 |

| Location | 1,072,845 – 1,072,919 |
|---|---|
| Length | 74 |
| Sequences | 10 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 55.67 |
| Shannon entropy | 0.90202 |
| G+C content | 0.61018 |
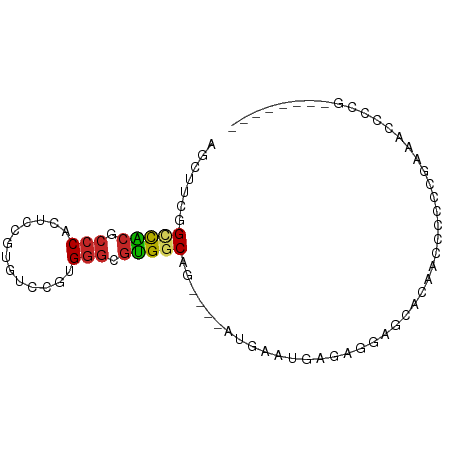
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -10.87 |
| Energy contribution | -10.65 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
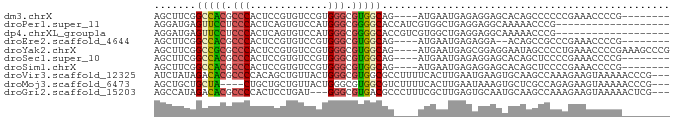
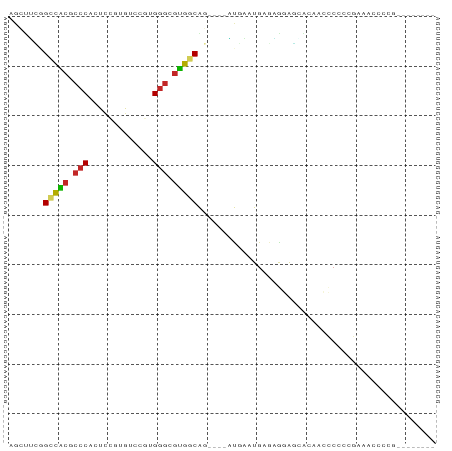
>dm3.chrX 1072845 74 + 22422827 AGCUUCGGCCACGCCCACUCCGUGUCCGUGGGCGUGGCAG----AUGAAUGAGAGGAGCACAGCCCCCCGAAACCCCG-------- .(((((.(((((((((((.........)))))))))))..----..........)))))...................-------- ( -29.32, z-score = -2.21, R) >droPer1.super_11 1229502 67 - 2846995 AGGAUGAGUUCCUCCCACUCAGUGUCCAUGGGCGGGGCACCAUCGUGGCUGAGGAGGCAAAAACCCG------------------- .((.......(((((...((((...((((((..((....)).))))))))))))))).......)).------------------- ( -21.54, z-score = 0.39, R) >dp4.chrXL_group1a 4123289 67 - 9151740 AGGAUGAGUUCCUCCCACUCAGUGUCCAUGGGCGGGGCACCGUCGUGGCUGAGGAGGCAAAAACCCG------------------- .((.......(((((...((((...((((((.(((....)))))))))))))))))).......)).------------------- ( -24.04, z-score = -0.21, R) >droEre2.scaffold_4644 1058016 72 + 2521924 AGCUUCGGCCACGCCCACUCCGUGUCCGUGGGCGUGGCAG----AUGAAUGAGAGGA--ACAGCCGCCCGAAACCCCG-------- ...(((((((((((((((.........)))))))))))..----..........((.--....))...))))......-------- ( -28.40, z-score = -1.85, R) >droYak2.chrX 986911 82 + 21770863 AGCUUCGGCCGCGCCCACUCCGUGUCCGUGGGCGUGGCAG----AUGAAUGAGCGGAGGAAUAGCCCCUGAAACCCCGAAAGCCCG .(((((((((((((((((.........)))))))))))..----.........(((.((......)))))......)).))))... ( -33.10, z-score = -2.06, R) >droSec1.super_10 903452 74 + 3036183 AGCUUCGGCCACGCCCACUCCGUGUCCGUGGGCGUGGCAG----AUGAAUGAGAGGAGCACAGCUCCCCGAAACCCCG-------- ...(((((((((((((((.........)))))))))))..----..........(((((...))))).))))......-------- ( -34.00, z-score = -3.47, R) >droSim1.chrX 863103 74 + 17042790 AGCUUCGGCCACGCCCACUCCGUGUCCGUGGGCGUGGCAG----AUGAAUGAGAGGAGCACAGCUCCCCGAAACCCCG-------- ...(((((((((((((((.........)))))))))))..----..........(((((...))))).))))......-------- ( -34.00, z-score = -3.47, R) >droVir3.scaffold_12325 42339 83 + 54264 AUCUAUAGACACGCCCCACAGCUGUUACUGGGCGUGGCGCCUUUUCACUUGAAUGAAGUGCAAGCCAAAGAAGUAAAAACCCG--- .(((.....(((((((.((....))....)))))))((((...(((....)))....)))).......)))............--- ( -19.70, z-score = -0.67, R) >droMoj3.scaffold_6473 7102590 79 + 16943266 AGCUGCUGCUA----CUGCUGCUGUUACUGGGCGUGGCGUCUUUUCACUUGAAUAAAGUGCUCGCCAGAGAAGUAAAAACCCG--- (((.((.....----..)).))).(((((.((((.((((.((((((....))..)))))))))))).....))))).......--- ( -20.40, z-score = 0.09, R) >droGri2.scaffold_15203 4234580 80 - 11997470 AGCCAUAGACACGCCCCACUCCUGAU---GGGCGUGACGCCCUUUCGCUUGAGUGCAAUGCAAGCCAAAGAAGUAAAAACUCG--- .........(((((((..(....)..---)))))))((...((((.(((((.........))))).))))..)).........--- ( -20.50, z-score = -1.22, R) >consensus AGCUUCGGCCACGCCCACUCCGUGUCCGUGGGCGUGGCAG____AUGAAUGAGAGGAGCACAACCCCCCGAAACCCCG________ .......(((((.(((.............))).)))))................................................ (-10.87 = -10.65 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:13 2011