| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,549,572 – 9,549,690 |
| Length | 118 |
| Max. P | 0.677444 |

| Location | 9,549,572 – 9,549,690 |
|---|---|
| Length | 118 |
| Sequences | 14 |
| Columns | 126 |
| Reading direction | forward |
| Mean pairwise identity | 69.95 |
| Shannon entropy | 0.66140 |
| G+C content | 0.49172 |
| Mean single sequence MFE | -30.01 |
| Consensus MFE | -12.89 |
| Energy contribution | -13.41 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.677444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
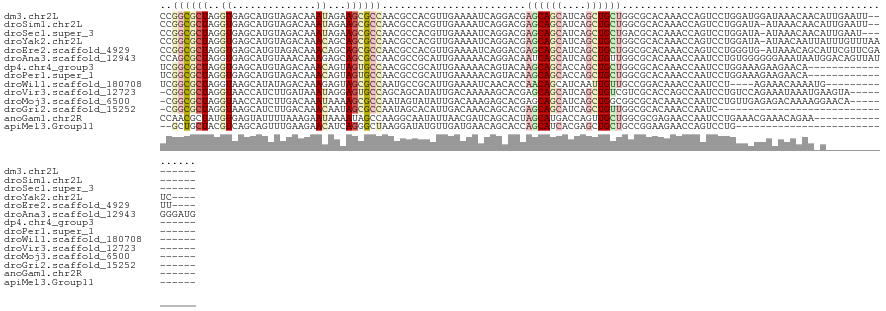
>dm3.chr2L 9549572 118 + 23011544 CCGGCGCUAGGUGAGCAUGUAGACAAAUAGAAGCGCCAACGCCACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGGCGCACAAACCAGUCCUGGAUGGAUAAACAACAUUGAAUU-------- ((((((...((((....(((......)))....))))..))))..........(((((((.((((((....))))))((........)).)))))))..)).................-------- ( -35.80, z-score = -1.62, R) >droSim1.chr2L 9321541 117 + 22036055 CCGGCGCUAGGUGAGCAUGUAGACAAAUAGAAGCGCCAACGCCACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGGCGCACAAACCAGUCCUGGAUA-AUAAACAACAUUGAAUU-------- ..((((...((((....(((......)))....))))..))))..((((....(((((((.((((((....))))))((........)).)))))))...-.....))))........-------- ( -35.50, z-score = -2.19, R) >droSec1.super_3 4996866 116 + 7220098 CCGGCGCUAGGUGAGCAUGUAGACAAAUAGAAGCGCCAACGCCACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGACGCACAAACCAGUCCUGGAUA-AUAAACAACAUUGAAU--------- ..((((...((((....(((......)))....))))..))))..((((....(((((((.((((((....))))))(.....)......)))))))...-.....)))).......--------- ( -32.20, z-score = -1.79, R) >droYak2.chr2L 12225541 121 + 22324452 CCGGCGCUAGGUGAGCAUGUAGACAAACAGCAGCGCCAACGCCACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGGCGCACAAACCAGUCCUGGAUA-AUAACAAUUAUUUGUUUAAUC---- ..((((...((((.((.(((......)))))..))))..))))..((((....(((((((.((((((....))))))((........)).)))))))...-.))))................---- ( -38.40, z-score = -2.29, R) >droEre2.scaffold_4929 10155003 121 + 26641161 CCGGCGCUAGGUGAGCAUGUAGACAAACAGCAGCGCCAACGCCACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGGCGCACAAACCAGUCCUGGGUG-AUAAACAGCAUUCGUUCGAUU---- ..((((...((((.((.(((......)))))..))))..))))..((((((..(((((((.((((((....))))))((........)).)))))))((.-....))........)))))).---- ( -39.50, z-score = -1.03, R) >droAna3.scaffold_12943 923427 126 + 5039921 CCAGCGCUAGGUGAGCAUGUAAACAAAGAGCAGCGCCAACGCCGCAUUGAAAAACAGGACAAUCAGCAUCAGCUGUUGGCGCACAAACCAAUCCUGUGGGGGGAAAUAAUGGACAGUUAUGGGAUG ...(((((.....))).((....))....)).(((((((((((...((....))..)).)....(((....)))))))))))........(((((((((..(...........)..))))))))). ( -31.40, z-score = 0.87, R) >dp4.chr4_group3 11064779 108 - 11692001 UCGGCGCUAGGUGAGCAUGUAGACAAACAGUAGUGCCAACGCCGCAUUGAAAAACAGUACAAGCAGCACCAGCUGCUGGCGCACAAACCAAUCCUGGAAAGAAGAACA------------------ ..(((((((.((...(.....)....))..)))))))..(((((.((((.....)))).).((((((....))))))))))......(((....)))...........------------------ ( -30.30, z-score = -0.90, R) >droPer1.super_1 8179828 108 - 10282868 UCGGCGCUAGGUGAGCAUGUAGACAAACAGUAGUGCCAACGCCGCAUUGAAAAACAGUACAAGCAGCACCAGCUGCUGGCGCACAAACCAAUCCUGGAAAGAAGAACA------------------ ..(((((((.((...(.....)....))..)))))))..(((((.((((.....)))).).((((((....))))))))))......(((....)))...........------------------ ( -30.30, z-score = -0.90, R) >droWil1.scaffold_180708 335929 107 - 12563649 UCGGCGCUAGGUAAGCAUAUAGACAAAGAGUAGCGCCAAUGCCGCAUUGAAAAUCAACACCAACAGCAUCAAUUGUUGCCGGACAAACCAAUCCU----AGAAACAAAAUG--------------- ..(((((((.....................)))))))..(((((..(((.....)))......(((((.....))))).))).))..........----............--------------- ( -18.40, z-score = -0.01, R) >droVir3.scaffold_12723 4017745 114 + 5802038 -CGGCGCUAGGUAACCAUCUUGAUAAAUAGGAGUGCCAGCAGCAUAUUGACAAAAAGCACGAGCAGCAUCAGCUGUCGUCGCACCAGCCAAUCCUGUCCAGAAAUAAAUGAAGUA----------- -.((((((.((((....(((((.....))))).)))))))................((((((.((((....)))))))).))....)))..........................----------- ( -28.90, z-score = -1.07, R) >droMoj3.scaffold_6500 17645589 114 - 32352404 -CGGCGCUAGGUAACCAUCUUGACAAAUAAAAGCGCCAAUAGUAUAUUGACAAAGAGCACGAGCAGCAUCAGCUGGCGGCGCACAAACCAAUCCUGUUGAGAGACAAAAGGAACA----------- -.((((((((((....))))...........)))))).................(.((.((..((((....)))).)))).).........(((((((....)))...))))...----------- ( -26.20, z-score = -0.54, R) >droGri2.scaffold_15252 13551589 92 - 17193109 -CGGCGCUAGGUAAGCAUCUUGACAAACAAUAGCGCCAAUAGCACAUUGACAAACAGCACGAGCAGCAUCAGCUGUUGGCGCACAAACCAAUC--------------------------------- -.((.(((.....)))................(((((((((((....((.....))((....)).......))))))))))).....))....--------------------------------- ( -25.90, z-score = -2.00, R) >anoGam1.chr2R 46119206 109 - 62725911 CCAACGCUAUGUGAGUAUUUUAAAGAAUAAAAUAGCCAAGGCAAUAUUAACGAUCAGCACUAGCAUGACCAGUUGCUGGCGCGAGAACCAAUCCUGAAACGAAACAGAA----------------- .....(((.((.(..(((((((.....))))))).))).)))...........((.((.((((((........)))))).))..)).......(((........)))..----------------- ( -20.00, z-score = -0.64, R) >apiMel3.Group11 3922956 94 + 12576330 --GCUGCUACGUCAGCAGUUUGAAGAACAUCAGGGCUAAGGAUAUGUUGAUGAACAGCACCAGCAUCACGAGCUGCUGCCGGAAGAACCAGUCCUG------------------------------ --((((((.....))))))...........(((((((..((...((((....))))(((.((((.......)))).)))(....)..)))))))))------------------------------ ( -27.30, z-score = -0.52, R) >consensus CCGGCGCUAGGUGAGCAUGUAGACAAACAGAAGCGCCAACGCCACAUUGAAAAACAGCACGAGCAGCAUCAGCUGCUGGCGCACAAACCAAUCCUGGAUAGAAAAACAA_A_______________ ..((((((..((..............))...))))))........................((((((....))))))................................................. (-12.89 = -13.41 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:31 2011