
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,031,363 – 1,031,454 |
| Length | 91 |
| Max. P | 0.841694 |

| Location | 1,031,363 – 1,031,454 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.31 |
| Shannon entropy | 0.35651 |
| G+C content | 0.45087 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -16.42 |
| Energy contribution | -16.49 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
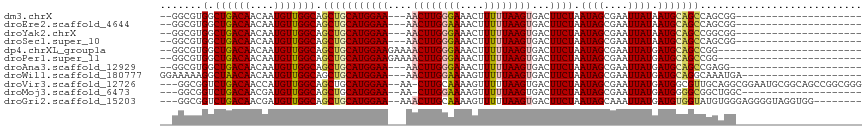
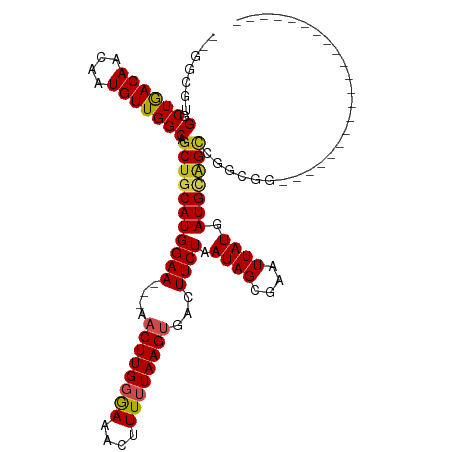
>dm3.chrX 1031363 91 + 22422827 --GGCGUGGCUGACAACAAUGUUGGCAGCUGCAUGGAA---AACUUGGGAAACUUUUUAAGUGACUUCUAAUAGCGAAUUAUAAUGCAGCCAGCGG--------------------- --((((..((((.((((...)))).))))..).(((((---.((((((((....))))))))...)))))...(((........))).))).....--------------------- ( -26.50, z-score = -1.83, R) >droEre2.scaffold_4644 1013819 91 + 2521924 --GGCGUGGCUGACAACAAUGUUGGCAGCUGCAUGGAA---AACUUGGAAAACUUUUUAAGUGACUUCUAAUAGCGAAUUAUAAUGCAGCCAGCGG--------------------- --((((..((((.((((...)))).))))..).(((((---.((((((((....))))))))...)))))...(((........))).))).....--------------------- ( -26.90, z-score = -2.14, R) >droYak2.chrX 944408 91 + 21770863 --GGCGUGGCUGACAACAAUGUUGGCAGCUGCAUGGAA---AACUUGGGAAACUUUUUAAGUGACUUCUAAUAGCGAAUUAUAAUGCAGCCGGCGG--------------------- --((((..((((.((((...)))).))))..).(((((---.((((((((....))))))))...)))))...(((........))).))).....--------------------- ( -26.50, z-score = -1.86, R) >droSec1.super_10 858254 91 + 3036183 --GGCGUGGCUGACAACAAUGUUGGCAGCUGCAUGGAA---AACUUGGGAAACUUUUUAAGUGACUUCUAAUAGCGAAUUAUAAUGCAGCCAGCGG--------------------- --((((..((((.((((...)))).))))..).(((((---.((((((((....))))))))...)))))...(((........))).))).....--------------------- ( -26.50, z-score = -1.83, R) >dp4.chrXL_group1a 4064066 91 - 9151740 --GGCGUGGCUGACAACAAUGUUGGCAGCUGCAUGGAAGAAAACUUGGGAAACUUUUUAAGUGACUUCUAAUAGCGAAUUAUGAUGCAGCCGG------------------------ --((((..((((.((((...)))).))))..).((((((...((((((((....))))))))..))))))...(((........))).)))..------------------------ ( -29.70, z-score = -3.38, R) >droPer1.super_11 1171226 91 - 2846995 --GGCGUGGCUGACAACAAUGUUGGCAGCUGCAUGGAAGAAAACUUGGGAAACUUUUUAAGUGACUUCUAAUAGCGAAUUAUGAUGCAGCCGG------------------------ --((((..((((.((((...)))).))))..).((((((...((((((((....))))))))..))))))...(((........))).)))..------------------------ ( -29.70, z-score = -3.38, R) >droAna3.scaffold_12929 829564 90 + 3277472 --GGCGUGGCUGACAACAAUGUUGGCAGCUGCAUGGAA---AACUUGGGAAACUUUUUAAGUGACUUCUAAUAGCGAAUUAUGAUGCAGCCGAGG---------------------- --((((..((((.((((...)))).))))..).(((((---.((((((((....))))))))...)))))...(((........))).)))....---------------------- ( -26.50, z-score = -2.20, R) >droWil1.scaffold_180777 1304110 94 + 4753960 GGAAAAAGGCUAACAACAAUGUUGGCAGCUGCAUGGAA---AACUUGGAAAAGUUUUUAAGUGACUUCUAAUAGCGAAUUAUGAUGCAGGCAAAUGA-------------------- ........(((((((....)))))))..((((((.(((---(((((....))))))))..((((.(((.......))))))).))))))........-------------------- ( -23.80, z-score = -2.57, R) >droVir3.scaffold_12726 1321024 111 - 2840439 ---GGCGGUCUGACAACCAUGUUGGCAGCUGCAUGGAA--AA-CUUGCAAAAGUUUUUAAGUGACUUCUAAUAGCGAAUUAUGAUGGCGUUGCAGGCGGAAUGCGGCAGCCGGCGGG ---...(((......))).(((((((.(((((((.(((--((-(((....))))))))..(..((.((..((((....))))...)).))..).......))))))).))))))).. ( -34.00, z-score = -0.23, R) >droMoj3.scaffold_6473 7018839 91 + 16943266 ---GGCGGUCUGACAACGAUGUUGGCAGCUGCAUGGAA--AA-CUUGGAAAAGUUUUUAAGUGACUUCUAAUAGCGAAUUAUGAUGGGGCGGCUGGC-------------------- ---(((.((((.(((.((.((((((.((..((...(((--((-(((....))))))))..))..)).)))))).)).....)).).)))).)))...-------------------- ( -24.90, z-score = -1.78, R) >droGri2.scaffold_15203 4175086 104 - 11997470 ---GGCGGUCUGACAACGAUGUUGGCAGCUGCAUGGAA--AAACUUGCAAAAGUUUUUAAGUGACUUCUAAUAGCAAAUUAUGAUGUGGUAUGUGGGAGGGGUAGGUGG-------- ---.((((.(((.((((...)))).)))))))((..((--((((((....))))))))..)).((((((....(((.(((((...))))).)))...))))))......-------- ( -25.40, z-score = -1.14, R) >consensus __GGCGUGGCUGACAACAAUGUUGGCAGCUGCAUGGAA___AACUUGGGAAACUUUUUAAGUGACUUCUAAUAGCGAAUUAUGAUGCAGCCGGCGG_____________________ .......(.((((((....))))))).(((((((((((....((((((((....))))))))...)))).((((....)))).)))))))........................... (-16.42 = -16.49 + 0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:07 2011