
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,030,018 – 1,030,125 |
| Length | 107 |
| Max. P | 0.852437 |

| Location | 1,030,018 – 1,030,125 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.82 |
| Shannon entropy | 0.43052 |
| G+C content | 0.44486 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
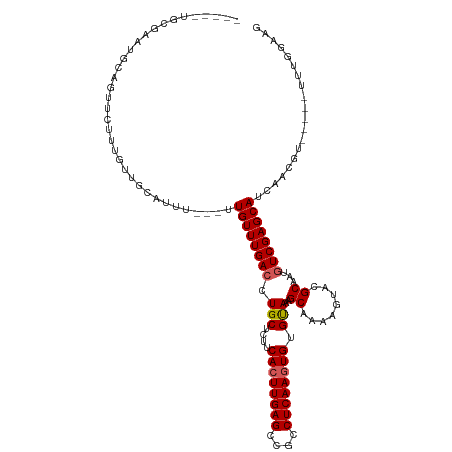
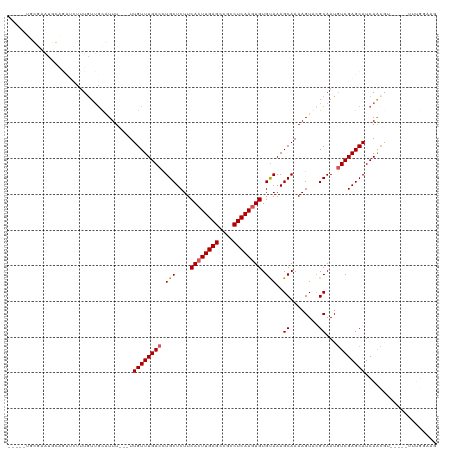
>dm3.chrX 1030018 107 + 22422827 -----UGCGUAUGGAGUUCUUUGUUGCGUUU---UUGUUUGACCUGCUCUUCACUUGAGCAGCCUCAAGUGUGUAAAGCAAAAGUGCGCAAUGUCGAGCAUCAACGU-----UUUGGAAG -----.((((.((..((((..((((((((((---((((((....(((....((((((((....)))))))).)))))))))))..))))))))..))))..))))))-----........ ( -34.50, z-score = -1.94, R) >droEre2.scaffold_4644 1012411 96 + 2521924 ---------------------UAUGUCAUUU---UUGUUUGACCUGCUCUUCACUUGAGGCGCCUCAAGUGUGUAAAGCAAAAGUACGCAAUGUCGAGCAUCAACAUCAACGUUUGGAAG ---------------------.((((.....---.((((((((.(((....((((((((....)))))))).)))..((........))...))))))))...))))............. ( -23.70, z-score = -0.86, R) >droYak2.chrX 942525 104 + 21770863 UGCGUUUGGAGUUCAGUUCUUUGUUGUAUUUGUGUUGUUUGACCUGCUCUUCAGUUGAGGCGCCUCAAGUGUGCAAAGCAAAAGUACGCAAUCUCGAGCAUCAG---------------- ...(((((((...(((....)))......((((((..((((...(((....((.(((((....))))).)).)))...))))...)))))))).))))).....---------------- ( -25.30, z-score = 0.34, R) >droSec1.super_10 856838 98 + 3036183 UGCGUAUGGAGUUC-----UUUGUUGCCUUU---UUGUUUGACUUGCUCUUCACUUGAGCCGCCUCAAGUGUGUAAAGCAAAAGUUAGCAAUGUCGAGCAUCAACG-------------- ..(((.((..((((-----..((((((.(((---((((((....(((....((((((((....)))))))).))))))))))))...))))))..))))..)))))-------------- ( -28.50, z-score = -1.92, R) >droSim1.chrX 819086 107 + 17042790 -----UGCGUAUGGAGUUCUUUGUUGCGUUU---UUGUUUGACCUGCUCUUCACUUGAGCCGCCUCAAGUGUGUAAAGCAAAAGUGCGCAAUGUCGAGCAUCAACGU-----UUUGGAAG -----.((((.((..((((..((((((((((---((((((....(((....((((((((....)))))))).)))))))))))..))))))))..))))..))))))-----........ ( -34.50, z-score = -2.12, R) >consensus _____UGCGAAUGCAGUUCUUUGUUGCAUUU___UUGUUUGACCUGCUCUUCACUUGAGCCGCCUCAAGUGUGUAAAGCAAAAGUACGCAAUGUCGAGCAUCAACGU_____UUUGGAAG ...................................((((((((.(((....((((((((....)))))))).)))..((........))...)))))))).................... (-20.78 = -21.02 + 0.24)

| Location | 1,030,018 – 1,030,125 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.82 |
| Shannon entropy | 0.43052 |
| G+C content | 0.44486 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
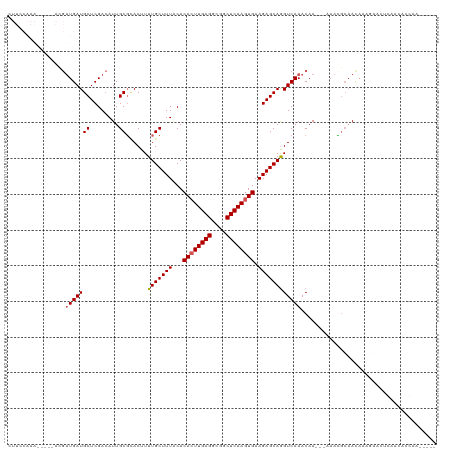
>dm3.chrX 1030018 107 - 22422827 CUUCCAAA-----ACGUUGAUGCUCGACAUUGCGCACUUUUGCUUUACACACUUGAGGCUGCUCAAGUGAAGAGCAGGUCAAACAA---AAACGCAACAAAGAACUCCAUACGCA----- ........-----.(((((..(.((....(((((......((((((...((((((((....)))))))).))))))(......)..---...)))))....)).)..)).)))..----- ( -24.10, z-score = -0.54, R) >droEre2.scaffold_4644 1012411 96 - 2521924 CUUCCAAACGUUGAUGUUGAUGCUCGACAUUGCGUACUUUUGCUUUACACACUUGAGGCGCCUCAAGUGAAGAGCAGGUCAAACAA---AAAUGACAUA--------------------- .......((((.(((((((.....)))))))))))....(((((((...((((((((....)))))))).)))))))((((.....---...))))...--------------------- ( -30.90, z-score = -3.35, R) >droYak2.chrX 942525 104 - 21770863 ----------------CUGAUGCUCGAGAUUGCGUACUUUUGCUUUGCACACUUGAGGCGCCUCAACUGAAGAGCAGGUCAAACAACACAAAUACAACAAAGAACUGAACUCCAAACGCA ----------------..............(((((...((((((((...((.(((((....))))).)).))))))))...................((......))........))))) ( -18.30, z-score = 0.78, R) >droSec1.super_10 856838 98 - 3036183 --------------CGUUGAUGCUCGACAUUGCUAACUUUUGCUUUACACACUUGAGGCGGCUCAAGUGAAGAGCAAGUCAAACAA---AAAGGCAACAAA-----GAACUCCAUACGCA --------------.((((.....))))..(((.....((((((((...((((((((....)))))))).))))))))........---...(....)...-----...........))) ( -24.30, z-score = -1.16, R) >droSim1.chrX 819086 107 - 17042790 CUUCCAAA-----ACGUUGAUGCUCGACAUUGCGCACUUUUGCUUUACACACUUGAGGCGGCUCAAGUGAAGAGCAGGUCAAACAA---AAACGCAACAAAGAACUCCAUACGCA----- ........-----.(((((..(.((....(((((......((((((...((((((((....)))))))).))))))(......)..---...)))))....)).)..)).)))..----- ( -24.10, z-score = -0.43, R) >consensus CUUCCAAA_____ACGUUGAUGCUCGACAUUGCGCACUUUUGCUUUACACACUUGAGGCGGCUCAAGUGAAGAGCAGGUCAAACAA___AAACGCAACAAAGAACUCAACACCAA_____ ................(((((((........))......(((((((...((((((((....)))))))).))))))))))))...................................... (-20.98 = -21.22 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:06 2011