| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,013,352 – 1,013,485 |
| Length | 133 |
| Max. P | 0.971906 |

| Location | 1,013,352 – 1,013,485 |
|---|---|
| Length | 133 |
| Sequences | 4 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 67.48 |
| Shannon entropy | 0.53464 |
| G+C content | 0.41084 |
| Mean single sequence MFE | -41.16 |
| Consensus MFE | -17.38 |
| Energy contribution | -18.88 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
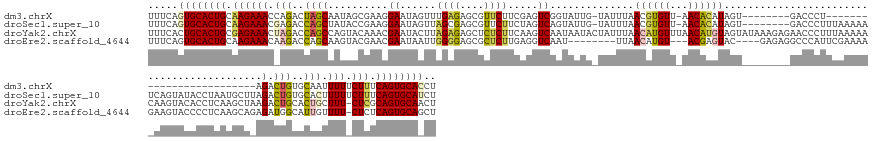
>dm3.chrX 1013352 133 + 22422827 UUUCAGUGCACUGCAAGAAACCAGACUAGCAAUAGCGAAGGAAUAGUUUGAGAGCGUUCUUCGAGUCGGUAUUG-UAUUUAACGUGUU-AACACAUAGU--------GACCCU-------------------------AGACUGUGCAAUUUUUCUUUCAGUGCACCU .....((((((((.((((((((.((((.((....))((((((...(((....))).)))))).)))))))((((-(.......(((..-..)))(((((--------......-------------------------..))))))))))..))))).)))))))).. ( -40.90, z-score = -3.04, R) >droSec1.super_10 840609 158 + 3036183 UUUCAGUGCACUGCAAGAAACGAGACCAGCUAUACCGAAGGAAUAGUUAGCGAGCGUUCUUCUAGUCAGUAUUG-UAUUUAACGUGUU-AACACAUAGU--------GACCCUUUAAAAAUCAGUAUACCUAAUGCUUAGACUGUGCACUUUUUCUUUCAGUGCAUCU .....((((((((.((((((.(((............((((((...(((....))).))))))((((((((((((-........(((..-..)))(((.(--------((...........))).)))...)))))))..)))))....))))))))).)))))))).. ( -37.90, z-score = -1.35, R) >droYak2.chrX 925714 167 + 21770863 UUUCACUGCACUGCGAGAAACUAGACCAGCCAGUACAAACGAAUACUUAGAGAGCUCUCUUCAAGUCAAUAAUACUAUUUAACAUGUUUAACAUGUAGUAUAAAGAGAACCCUUUAAAAACAAGUACACCUCAAGCUAAGACUGCACUGCUUU-CUCGCAGUGCAACU ......((((((((((((((.(((........((((........((((..(((....)))..))))......((((.....(((((.....)))))))))(((((......))))).......)))).......((.......)).))).)))-)))))))))))... ( -45.00, z-score = -4.30, R) >droEre2.scaffold_4644 995921 152 + 2521924 UUUCAGUGCACUGCAAGAAACAAGACCAGCAAGUACGAACGAAUAAUUGGGGAGCGCUCUUGAGGUCAAU--------UUAACAUGU---ACGAGUAC----GAGAGGCCCAUUCGAAAAGAAGUACCCCUCAAGCAGAGAUGGCAUUGUUUU-CUCUCAGUGCAGCU ......(((((((..((((((((..(((....((((..............((.((.((((((.(.((...--------.........---..)).).)----))))))))).(((.....)))))))..(((.....))).)))..))).)))-))..)))))))... ( -40.84, z-score = -0.81, R) >consensus UUUCAGUGCACUGCAAGAAACAAGACCAGCAAGAACGAACGAAUAGUUAGAGAGCGCUCUUCAAGUCAAUAUUG_UAUUUAACAUGUU_AACACAUAGU________GACCCUUUAAAAA_AAGUACACCUCAAGCU_AGACUGCACAGUUUU_CUCUCAGUGCAACU .....((((((((.((((((...((((........(....)........(((....)))....))))..............(((((.....))))).......................................................)))))).)))))))).. (-17.38 = -18.88 + 1.50)

| Location | 1,013,352 – 1,013,485 |
|---|---|
| Length | 133 |
| Sequences | 4 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 67.48 |
| Shannon entropy | 0.53464 |
| G+C content | 0.41084 |
| Mean single sequence MFE | -44.72 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
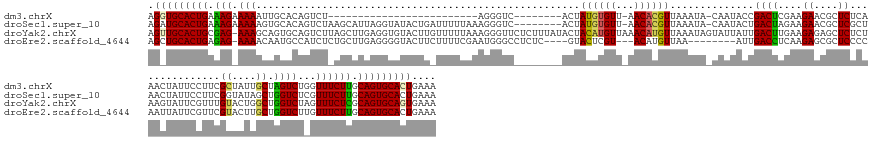
>dm3.chrX 1013352 133 - 22422827 AGGUGCACUGAAAGAAAAAUUGCACAGUCU-------------------------AGGGUC--------ACUAUGUGUU-AACACGUUAAAUA-CAAUACCGACUCGAAGAACGCUCUCAAACUAUUCCUUCGCUAUUGCUAGUCUGGUUUCUUGCAGUGCACUGAAA .(((((((((.((((((.......(((.((-------------------------((((((--------.....(((..-..)))((.....)-)......)))))((((..................))))((....))))).))).)))))).))))))))).... ( -37.07, z-score = -2.41, R) >droSec1.super_10 840609 158 - 3036183 AGAUGCACUGAAAGAAAAAGUGCACAGUCUAAGCAUUAGGUAUACUGAUUUUUAAAGGGUC--------ACUAUGUGUU-AACACGUUAAAUA-CAAUACUGACUAGAAGAACGCUCGCUAACUAUUCCUUCGGUAUAGCUGGUCUCGUUUCUUGCAGUGCACUGAAA ((.(((((((.((((((.(((((.........)))))((.(((((((((((((....((((--------(.((((((((-((....)).))))-).))).)))))..))))).(....)...........)))))))).)).......)))))).))))))))).... ( -38.50, z-score = -0.66, R) >droYak2.chrX 925714 167 - 21770863 AGUUGCACUGCGAG-AAAGCAGUGCAGUCUUAGCUUGAGGUGUACUUGUUUUUAAAGGGUUCUCUUUAUACUACAUGUUAAACAUGUUAAAUAGUAUUAUUGACUUGAAGAGAGCUCUCUAAGUAUUCGUUUGUACUGGCUGGUCUAGUUUCUCGCAGUGCAGUGAAA ..((((((((((((-(((..((..(((((...((..((.(.(((((((.......((((((((((((((((((((((.....))))).....))))).........)))))))))))).))))))).).)).))...)))))..))..)))))))))))))))..... ( -58.10, z-score = -3.99, R) >droEre2.scaffold_4644 995921 152 - 2521924 AGCUGCACUGAGAG-AAAACAAUGCCAUCUCUGCUUGAGGGGUACUUCUUUUCGAAUGGGCCUCUC----GUACUCGU---ACAUGUUAA--------AUUGACCUCAAGAGCGCUCCCCAAUUAUUCGUUCGUACUUGCUGGUCUUGUUUCUUGCAGUGCACUGAAA ((.(((((((..((-(((.(((.((((.........((((((..((((.....))).)..))))))----(((...((---((.......--------.(((....)))(((((.............))))))))).))))))).))))))))..))))))))).... ( -45.22, z-score = -2.00, R) >consensus AGAUGCACUGAAAG_AAAACAGCACAGUCU_AGCUUGAGGUGUACUU_UUUUUAAAGGGUC________ACUACGUGUU_AACACGUUAAAUA_CAAUACUGACUUGAAGAACGCUCUCUAACUAUUCCUUCGUAAUUGCUGGUCUAGUUUCUUGCAGUGCACUGAAA .(((((((((.((((((.......................................................(((((.....)))))..............((((....((....))...............((....)).))))...)))))).))))))))).... (-20.04 = -20.23 + 0.19)
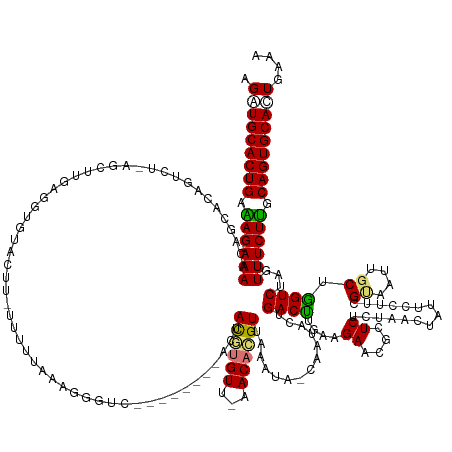
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:05:01 2011