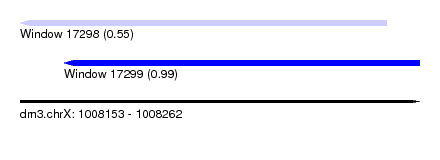
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,008,153 – 1,008,262 |
| Length | 109 |
| Max. P | 0.992568 |

| Location | 1,008,153 – 1,008,253 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.55794 |
| G+C content | 0.38792 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -7.63 |
| Energy contribution | -7.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1008153 100 - 22422827 ---UGGUCCACUUUCA-------AGCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGC-UCUCCAUGUGUGU---------- ---.....(((.....-------(((((.((((((.....))))))((.((((..((((((..............))))))..)))).))...))))-)......)))...---------- ( -22.04, z-score = -2.77, R) >droEre2.scaffold_4644 990560 100 - 2521924 ---UGGUCCACUUUCA-------AGCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGC-UCUCCAUGUGUGU---------- ---.....(((.....-------(((((.((((((.....))))))((.((((..((((((..............))))))..)))).))...))))-)......)))...---------- ( -22.04, z-score = -2.77, R) >droYak2.chrX 919961 100 - 21770863 ---UGGUCCACUUUCA-------AGCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGC-UCUCCAUGUGUGU---------- ---.....(((.....-------(((((.((((((.....))))))((.((((..((((((..............))))))..)))).))...))))-)......)))...---------- ( -22.04, z-score = -2.77, R) >droSec1.super_10 835383 100 - 3036183 ---UGGUCCACUUUCA-------AGCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGC-UCUCCAUGUGUGU---------- ---.....(((.....-------(((((.((((((.....))))))((.((((..((((((..............))))))..)))).))...))))-)......)))...---------- ( -22.04, z-score = -2.77, R) >droSim1.chrX_random 873127 100 - 5698898 ---UGGUCCACUUUCA-------AGCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGC-UCUCCAUGUGUGU---------- ---.....(((.....-------(((((.((((((.....))))))((.((((..((((((..............))))))..)))).))...))))-)......)))...---------- ( -22.04, z-score = -2.77, R) >droWil1.scaffold_180777 1270583 102 - 4753960 -------UGGCUUGCA-UCAUCCUUUUUGGACCACUUUUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUGCUUUUCGGGUAGAU----------- -------.....((((-(......(((((((((((.....))))))...(((((....((....)).....)))))))))).....))))).((((((.....)))))).----------- ( -19.60, z-score = -1.35, R) >droMoj3.scaffold_6473 6975964 117 - 16943266 ---UGGUCCAGCUCCAUUUCGAGAGUGGCGACCACUUUUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUGC-UUUCGGCUACUUUAUAUGGGUGU ---.......((.((((....((((((((((((((.....))))))((.((((..((((((..............))))))..)))).)).......-.....))))))))..)))))).. ( -30.54, z-score = -2.63, R) >droGri2.scaffold_15203 4134437 100 + 11997470 ---UGGUCCAGAUCCACUU------------CCACUUUUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUGC-UUUCGGCUACUUUAUUUA----- ---(((.......)))...------------.........(((((((..(((((....((....)).....)))))............((.....))-...)))))))........----- ( -13.50, z-score = -1.03, R) >droPer1.super_11 1136381 107 + 2846995 UGGUCUGCUGCCUGCA--CAUCCUGCACAUCCCACUUUUUGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUCCACUUCUGUCUGU------------ .((..(((.....)))--...))((((....((((.....)))).....(((((....((....)).....)))))...........))))..................------------ ( -14.80, z-score = -0.49, R) >dp4.chrXL_group1a 4029057 107 + 9151740 UGGUCUGCUGCCUGCA--CAUCCUGCACAUCCCACUUUUUGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUCCACUUCUGUCUGU------------ .((..(((.....)))--...))((((....((((.....)))).....(((((....((....)).....)))))...........))))..................------------ ( -14.80, z-score = -0.49, R) >droAna3.scaffold_12929 798866 79 - 3277472 -----------------------------UGGCAAGUGGC-UAGUGGCUUCAAGUUUUUAUCCUUAUUUAUCCUUGUGAAAUAUUCAUGCACUCGGCUGCCAGGACUCU------------ -----------------------------(((((....((-(((((.....(((........)))..........((((.....)))).)))).)))))))).......------------ ( -13.00, z-score = 1.65, R) >droVir3.scaffold_12726 1282508 114 + 2840439 ---UGGCCCAGGUCCAUUUCG-GAGCGGCGACCACUUUUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUGC-UUUCGGCUACUUUAUAUGUGU-- ---(((((..(.(((.....)-)).)(((((((((.....))))))((.((((..((((((..............))))))..)))).)).....))-)...)))))............-- ( -27.64, z-score = -1.92, R) >consensus ___UGGUCCACUUUCA_______AGCAGCGACCACUUUUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGC_UCUCCGUGUGUGU__________ ...............................((((.....))))..((.((((..((((((..............))))))..)))).))............................... ( -7.63 = -7.73 + 0.11)



| Location | 1,008,165 – 1,008,262 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Shannon entropy | 0.50386 |
| G+C content | 0.39763 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -12.62 |
| Energy contribution | -13.06 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1008165 97 - 22422827 CUCUGCUGUUGGUC--CACUUUC-AA----GCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGCU------- ....((((((((..--.....))-.)----)))))((((((.....))))))((.((((..((((((..............))))))..)))).))........------- ( -21.74, z-score = -2.96, R) >droEre2.scaffold_4644 990572 97 - 2521924 GUCUGCUGUUGGUC--CACUUUC-AA----GCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGCU------- ....((((((((..--.....))-.)----)))))((((((.....))))))((.((((..((((((..............))))))..)))).))........------- ( -21.74, z-score = -2.79, R) >droYak2.chrX 919973 97 - 21770863 CCCUGCUGCUGGUC--CACUUUC-AA----GCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGCU------- ....((((((((..--.....))-.)----)))))((((((.....))))))((.((((..((((((..............))))))..)))).))........------- ( -24.14, z-score = -3.71, R) >droSec1.super_10 835395 97 - 3036183 CACUGCUGUUGGUC--CACUUUC-AA----GCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGCU------- ....((((((((..--.....))-.)----)))))((((((.....))))))((.((((..((((((..............))))))..)))).))........------- ( -21.74, z-score = -2.76, R) >droSim1.chrX_random 873139 97 - 5698898 CACUGCUGUUGGUC--CACUUUC-AA----GCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGCU------- ....((((((((..--.....))-.)----)))))((((((.....))))))((.((((..((((((..............))))))..)))).))........------- ( -21.74, z-score = -2.76, R) >droWil1.scaffold_180777 1270595 99 - 4753960 UUCUACUUUUGGCU--UGCAU---CAUCCUUUUUGGACCACUUUUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUGCU------- ..........(((.--(((((---......(((((((((((.....))))))...(((((....((....)).....)))))))))).....)))))....)))------- ( -18.60, z-score = -3.06, R) >droMoj3.scaffold_6473 6975979 103 - 16943266 --UUGGUCCAGCUC--CAUUUCGAGA----GUGGCGACCACUUUUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUGCUUUCGGCU --.(((.......)--))..((((((----(((((((((((.....)))))))))(((((....((....)).....)))))...................)))))))).. ( -25.70, z-score = -2.56, R) >droPer1.super_11 1136392 104 + 2846995 UCCUGCUGUGGUCUGCUGCCUGCACAUCCUGCACAUCCCACUUUUUGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUCCA------- ...(((.(.(((.(((.....))).)))).)))....((((.....))))..((.((((..((((((..............))))))..)))).))........------- ( -17.04, z-score = -1.18, R) >dp4.chrXL_group1a 4029068 104 + 9151740 UCCUGCUGUGGUCUGCUGCCUGCACAUCCUGCACAUCCCACUUUUUGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUCCA------- ...(((.(.(((.(((.....))).)))).)))....((((.....))))..((.((((..((((((..............))))))..)))).))........------- ( -17.04, z-score = -1.18, R) >droAna3.scaffold_12929 798877 74 - 3277472 -----------------------------GGCUGGUGGCAAGUGGC-UAGUGGCUUCAAGUUUUUAUCCUUAUUUAUCCUUGUGAAAUAUUCAUGCACUCGGCU------- -----------------------------((((((((......((.-(((..((.....))..))).))............((((.....)))).))).)))))------- ( -13.40, z-score = 1.20, R) >droVir3.scaffold_12726 1282521 104 + 2840439 CUUUGGCCCAGGUC--CAUUUCG-GA----GCGGCGACCACUUUUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUUUGCUUUCGGCU ....((((..(.((--(.....)-))----.)(((((((((.....))))))((.((((..((((((..............))))))..)))).)).....)))...)))) ( -26.34, z-score = -2.23, R) >consensus CUCUGCUGUUGGUC__CACUUUC_AA____GCAGCGACCACUUAUCGUGGUCGUUUGAAGUUUUUAUCCUUAUUUAUCUUCAUAAAAUAUUCAUGCACUCUGCU_______ ..............................((((.((((((.....))))))((.((((..((((((..............))))))..)))).))...))))........ (-12.62 = -13.06 + 0.44)

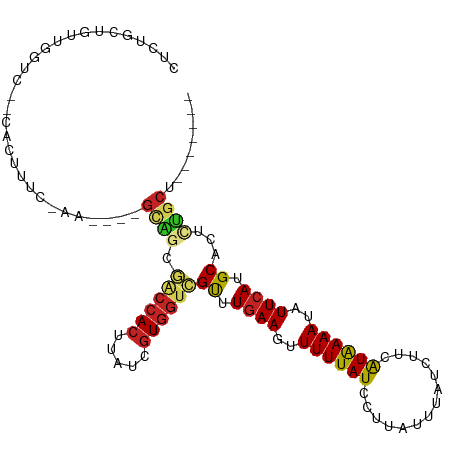
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:58 2011