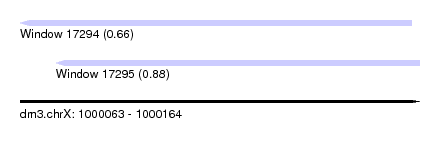
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,000,063 – 1,000,164 |
| Length | 101 |
| Max. P | 0.879201 |

| Location | 1,000,063 – 1,000,162 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Shannon entropy | 0.40879 |
| G+C content | 0.49763 |
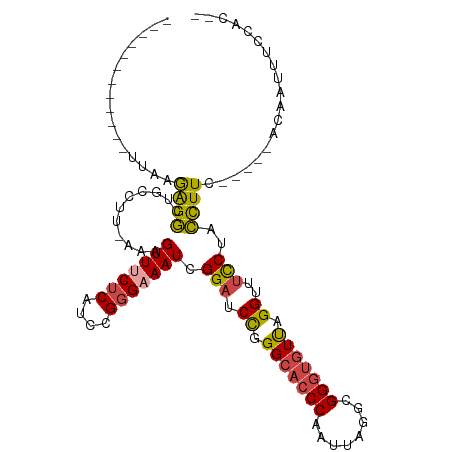
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -20.91 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
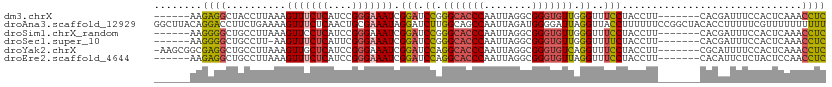
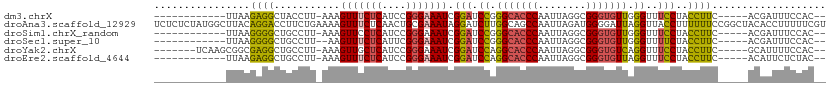
>dm3.chrX 1000063 99 - 22422827 ------AAGAGGCUACCUUAAAGUUUCUCAUCCGGGAAAUCGGAUCCGGGCACCCAAUUAGGCGGGUGUUGGGUUUCCUACCUU-------CACGAUUUCCACUCAAACCUC ------.(((((((.......)))))))......((((((((((.((.(((((((........))))))).))..........)-------).))))))))........... ( -30.90, z-score = -1.38, R) >droAna3.scaffold_12929 789018 112 - 3277472 GGCUUACAGGACCUUCUGAAAAGUUUCUCAACUGCGAAAUAGGAUCUUGGCAGCCAAUUAGAUGGGGAUUAGGUUACCUUUUUUCCGGCUACACCUUUUUCGUUUUUUUUUU ((((..((((((((.......((((....)))).......))).)))))..))))...(((.(((..(..(((...)))..)..))).)))..................... ( -27.24, z-score = -0.92, R) >droSim1.chrX_random 862803 99 - 5698898 ------AAGGGGCUGCCUUAAAGUUCCUCAUCCGGGAAAUCGGAUCCGGGCACCCAAUUAGGCGGGUGUUGGGUUUCCUACCUU-------CACGAUUUCCACUCAAACCUC ------.(((((((.......)))))))......((((((((((.((.(((((((........))))))).))..........)-------).))))))))........... ( -33.60, z-score = -1.12, R) >droSec1.super_10 827257 98 - 3036183 ------AAGGGGCUGCCUU-AAGUUUCUCAUUCGGGAAAUCGGAUCCGGGCACCCAAUUAGGCGGGUGUUGGGUUUUCUACCUU-------CACGAUUUCCACUCAAACCUC ------.((..(((.....-.)))..))......((((((((((.((.(((((((........))))))).))..........)-------).))))))))........... ( -32.30, z-score = -1.66, R) >droYak2.chrX 910539 104 - 21770863 -AAGCGGCGAGGCUGCCUUAAAGUUGCUCAUCCGGGAAAUCGGAUCCAGGCACCCAAUUAGGCGGGUGUCAGGUUUCCUACCUU-------CGCAUUUUCCACUCAAACCUC -.((((((((((...))))...)))))).....(((((((((((....(((((((........))))))).(((.....)))))-------)).)))))))........... ( -32.30, z-score = -1.00, R) >droEre2.scaffold_4644 977662 99 - 2521924 ------AAGAGGCUGCCUUAAAGUUUCUCAUCCGGGAAAUCGGAUCCAGGCACCCAAUUAGGCGGGUGUUAGGUUUCCUACCUU-------CACAUUCUCUACUCCAACCUC ------.((((..((.......(((((((....))))))).(((.((..((((((........))))))..))..)))......-------..))..))))........... ( -28.00, z-score = -1.31, R) >consensus ______AAGAGGCUGCCUUAAAGUUUCUCAUCCGGGAAAUCGGAUCCGGGCACCCAAUUAGGCGGGUGUUAGGUUUCCUACCUU_______CACGAUUUCCACUCAAACCUC ........((((..........(((((((....))))))).(((.((.(((((((........))))))).))..)))..............................)))) (-20.91 = -21.17 + 0.25)
| Location | 1,000,072 – 1,000,164 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.13 |
| Shannon entropy | 0.41444 |
| G+C content | 0.49855 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.25 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
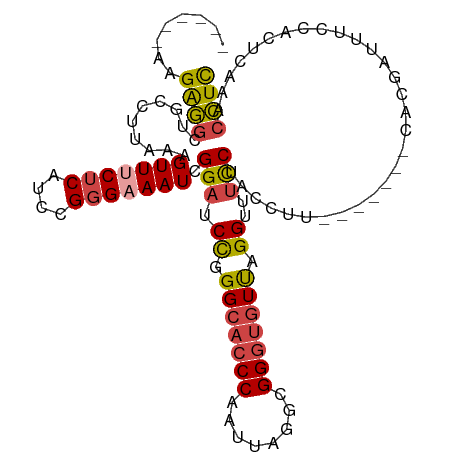
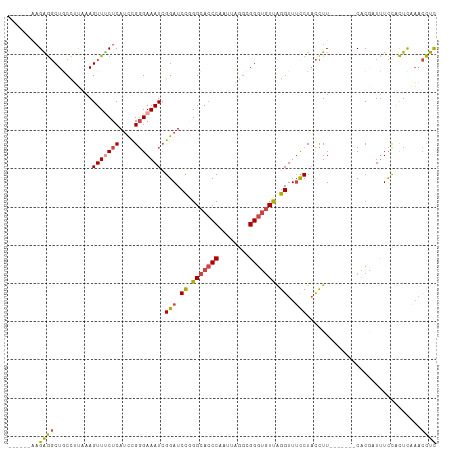
>dm3.chrX 1000072 92 - 22422827 ------------UUAAGAGGCUACCUU-AAAGUUUCUCAUCCGGGAAAUCGGAUCCGGGCACCCAAUUAGGCGGGUGUUGGGUUUCCUACCUUC-----ACGAUUUCCAC-- ------------...(((((((.....-..)))))))......((((((((((.((.(((((((........))))))).))..........))-----.))))))))..-- ( -30.90, z-score = -1.82, R) >droAna3.scaffold_12929 789027 112 - 3277472 UCUCUCUAUGGCUUACAGGACCUUCUGAAAAGUUUCUCAACUGCGAAAUAGGAUCUUGGCAGCCAAUUAGAUGGGGAUUAGGUUACCUUUUUUCCGGCUACACCUUUUUCGU .((.(((((((((..((((((((.......((((....)))).......))).)))))..)))))..)))).))(((..((((((((........)).)).))))..))).. ( -29.04, z-score = -0.92, R) >droSim1.chrX_random 862812 92 - 5698898 ------------UUAAGGGGCUGCCUU-AAAGUUCCUCAUCCGGGAAAUCGGAUCCGGGCACCCAAUUAGGCGGGUGUUGGGUUUCCUACCUUC-----ACGAUUUCCAC-- ------------...(((((((.....-..)))))))......((((((((((.((.(((((((........))))))).))..........))-----.))))))))..-- ( -33.60, z-score = -1.59, R) >droSec1.super_10 827266 91 - 3036183 ------------UUAAGGGGCUGCCUU--AAGUUUCUCAUUCGGGAAAUCGGAUCCGGGCACCCAAUUAGGCGGGUGUUGGGUUUUCUACCUUC-----ACGAUUUCCAC-- ------------...((..(((.....--.)))..))......((((((((((.((.(((((((........))))))).))..........))-----.))))))))..-- ( -32.30, z-score = -2.30, R) >droYak2.chrX 910548 97 - 21770863 -------UCAAGCGGCGAGGCUGCCUU-AAAGUUGCUCAUCCGGGAAAUCGGAUCCAGGCACCCAAUUAGGCGGGUGUCAGGUUUCCUACCUUC-----GCAUUUUCCAC-- -------...((((((((((...))))-...)))))).....(((((((((((....(((((((........))))))).(((.....))))))-----).)))))))..-- ( -32.30, z-score = -1.38, R) >droEre2.scaffold_4644 977671 92 - 2521924 ------------UUAAGAGGCUGCCUU-AAAGUUUCUCAUCCGGGAAAUCGGAUCCAGGCACCCAAUUAGGCGGGUGUUAGGUUUCCUACCUUC-----ACAUUCUCUAC-- ------------...((((..((....-...(((((((....))))))).(((.((..((((((........))))))..))..))).......-----.))..))))..-- ( -28.00, z-score = -1.90, R) >consensus ____________UUAAGAGGCUGCCUU_AAAGUUUCUCAUCCGGGAAAUCGGAUCCGGGCACCCAAUUAGGCGGGUGUUAGGUUUCCUACCUUC_____ACAAUUUCCAC__ ................((((...........(((((((....))))))).(((.((.(((((((........))))))).))..)))..))))................... (-20.30 = -20.25 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:54 2011