| Sequence ID | dm3.chrX |
|---|---|
| Location | 998,920 – 999,019 |
| Length | 99 |
| Max. P | 0.712322 |

| Location | 998,920 – 999,019 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.76 |
| Shannon entropy | 0.54563 |
| G+C content | 0.38767 |
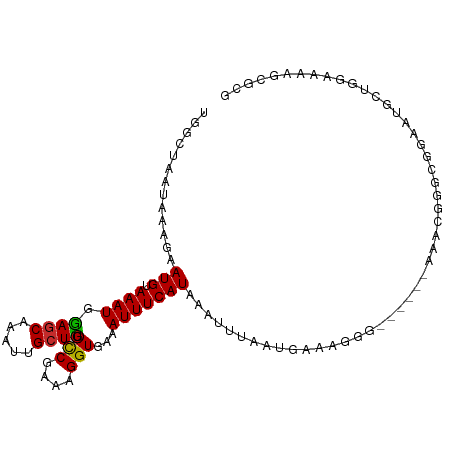
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -9.76 |
| Energy contribution | -9.50 |
| Covariance contribution | -0.26 |
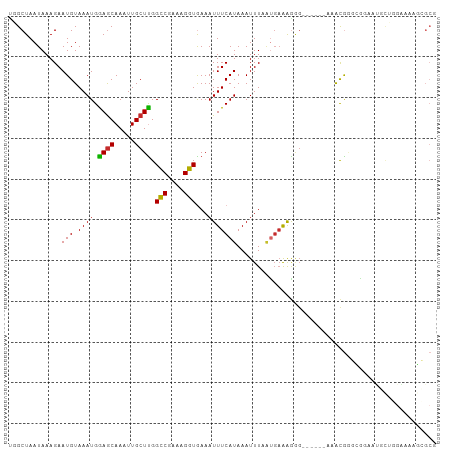
| Combinations/Pair | 1.21 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712322 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
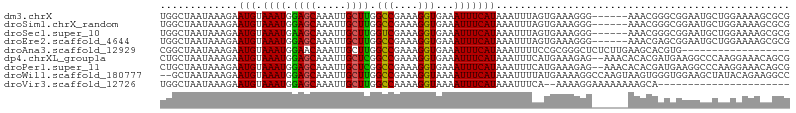
>dm3.chrX 998920 99 + 22422827 UGGCUAAUAAAGAAUGUAAAUGGAGCAAAUUGCUUGGCCGAAAGGUGAAAUUUCAUAAAUUUAGUGAAAGGG------AAACGGGCGGAAUGCUGGAAAAGCGCG ........................((...((((((((((....)))....((((((.......))))))...------...)))))))...(((.....))))). ( -21.70, z-score = -1.60, R) >droSim1.chrX_random 861676 99 + 5698898 UGGCUAAUAAAGAAUGUAAAUGGAGCAAAUUGCUUGGCCGAAAGGUGAAAUUUCAUAAAUUUAGUGAAAGGG------AAACGGGCGGAAUGCUGGAAAAGCGCG ........................((...((((((((((....)))....((((((.......))))))...------...)))))))...(((.....))))). ( -21.70, z-score = -1.60, R) >droSec1.super_10 826129 99 + 3036183 UGGCUAAUAAAGAAUGUAAAUGAAGCAAAUUGCUUGGUCGAAAGGUGAAAUUUCAUAAAUUUAGUGAAAGGG------AAACGGGCGGAAUGCUGGAAAAGCGCG ........................((...(((((((.((....)).....((((((.......))))))...------...)))))))...(((.....))))). ( -17.10, z-score = -0.49, R) >droEre2.scaffold_4644 976561 99 + 2521924 UGGCUAAUAAAGAAUGUAAAUGGAGCAAAUUGCUUGGCCGAAAGGUGAAAUUUCAUAAAUUUAGUGAAAGGG------AAACGAGCGGAAUGCUGGAAAAGCGCG ........................((...((((((((((....)))....((((((.......))))))...------...)))))))...(((.....))))). ( -22.50, z-score = -1.99, R) >droAna3.scaffold_12929 787519 87 + 3277472 CGGCUAAUAAAGAAUGUAAAUGGAACAAAUUGCUUGGCCGAAAGGUGAAAUUUCAUAAAUUUUCCGCGGGCUCUCUUGAAGCACGUG------------------ (((((....((((..((...((((((((.....)))(((....)))...............)))))...))..))))..))).))..------------------ ( -18.60, z-score = -0.46, R) >dp4.chrXL_group1a 4011612 103 - 9151740 CUGCUAAUAAAGAAUGUAAAUGGAGCAAAUUGCUCGGCCGAAAGGUGAAAUUUCAUAAAUUUCAUGAAAGAG--AAACACACGAUGAAGGCCCAAGGAAACAGCG ..(((...............((((((.....))))((((....((((...((((((.......))))))...--...))).)......)))))).(....)))). ( -22.20, z-score = -1.80, R) >droPer1.super_11 1122867 103 - 2846995 CUGCUAAUAAAGAAUGUAAAUGGAGCAAAUUGCUCGGCCGAAAGGUGAAAUUUCAUAAAUUUCAUGAAAGAG--AAACACACGAUGAAGGCCCAAGGAAACAGCG ..(((...............((((((.....))))((((....((((...((((((.......))))))...--...))).)......)))))).(....)))). ( -22.20, z-score = -1.80, R) >droWil1.scaffold_180777 3493404 103 - 4753960 --GCUAAUAAAGAAUGUAAAUGGAGCAAAUUGCUUGGCCGAAAGGUAAAAUUUCAUAAAUUUUAUGAAAAGGCCAAGUAAGUGGGUGGAAGCUAUACAGAAGGCC --(((........................((((((((((.....((((((((.....)))))))).....))))))))))((((((....))).)))....))). ( -23.90, z-score = -1.99, R) >droVir3.scaffold_12726 1265403 81 - 2840439 UGGCUAAUAAAGAAUGUAAAUGGAGCAAAUUGCUUGGCCAAAAGGUAAAAUUUCAUAAAUUUCA--AAAAGGAAAAAAAAGCA---------------------- ..(((........(((.((((.((((.....)))).(((....)))...)))))))...((((.--.....))))....))).---------------------- ( -10.90, z-score = -0.05, R) >consensus UGGCUAAUAAAGAAUGUAAAUGGAGCAAAUUGCUUGGCCGAAAGGUGAAAUUUCAUAAAUUUAAUGAAAGGG______AAACGGGCGGAAUGCUGGAAAAGCGCG .............(((.((((.((((.....)))).(((....)))...)))))))................................................. ( -9.76 = -9.50 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:52 2011