
| Sequence ID | dm3.chrX |
|---|---|
| Location | 995,270 – 995,384 |
| Length | 114 |
| Max. P | 0.878948 |

| Location | 995,270 – 995,381 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.86 |
| Shannon entropy | 0.26261 |
| G+C content | 0.44349 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -21.46 |
| Energy contribution | -24.65 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 995270 111 + 22422827 GAGAUGCCAGAUGCCAAUCUUGUAUCUUGUGGGUUGGGUUGGCUUCCCUAUUUUCGAGGAUUGCGAAUGCAUUUGGCAUUCUUUCCGCUAAUGACAAUCGCAAAUGUCUUG (((((((.((((....)))).)))))))(((((..((.....))..)))))...(((((.((((((.((((((.(((.........))))))).)).))))))...))))) ( -32.90, z-score = -1.35, R) >droEre2.scaffold_4644 973002 85 + 2521924 -------GAGAUGCCAAUCUUGU------------GGAUUGGUUUCCCUAUUUUCGGGGAUUG------CAUU-GCCAUUCUUUUCGCUAAUGACAAUCGCAAAUGUCUUG -------((((((((((((....------------.))))))).(((((......)))))(((------((((-(.((((.........)))).)))).))))..))))). ( -24.10, z-score = -2.09, R) >droSec1.super_10 822377 110 + 3036183 GAGAUACCAGAUGCCAAUCUUGUAUCUUGUGGGUUGGGUUGGCUUCCCUAUUUUCGGGGAUUGCAAAUGCAUU-GGCAUUCUUUCCGCUAAUGACAAUCGCAAAUGUCUUG ((((((......((((((((...(((.....))).)))))))).(((((......)))))((((...((((((-(((.........))))))).))...)))).)))))). ( -34.00, z-score = -1.89, R) >droSim1.chrX_random 857896 110 + 5698898 GAGAUGCCAGAUGCCAAUCUGGUAUCUUGUGGGUUGGGUUGGCUUCCCUAUUUUCGAAGAUUGCGAAUGCAUU-GGCAUUCUUUCCGCUAAUGACAAUCGCAAAUGUCUUG ((((((((((((....))))))))))))(((((..((.....))..))))).....((((((((((.((((((-(((.........))))))).)).)))))...))))). ( -42.10, z-score = -4.22, R) >consensus GAGAUGCCAGAUGCCAAUCUUGUAUCUUGUGGGUUGGGUUGGCUUCCCUAUUUUCGAGGAUUGCGAAUGCAUU_GGCAUUCUUUCCGCUAAUGACAAUCGCAAAUGUCUUG ((((((((((((....)))))))))))).........((((((.(((((......)))))....((((((.....)))))).....))))))((((........))))... (-21.46 = -24.65 + 3.19)

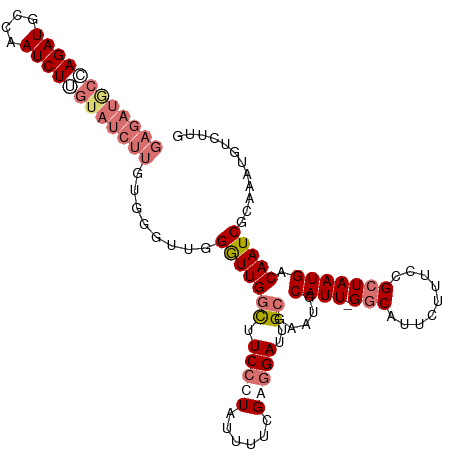

| Location | 995,273 – 995,384 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Shannon entropy | 0.25810 |
| G+C content | 0.44349 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 995273 111 + 22422827 AUGCCAGAUGCCAAUCUUGUAUCUUGUGGGUUGGGUUGGCUUCCCUAUUUUCGAGGAUUGCGAAUGCAUUUGGCAUUCUUUCCGCUAAUGACAAUCGCAAAUGUCUUGCGA (((((((((((.....((((((((((..(((.(((.......))).)))..)))))..)))))..)))))))))))......(((....((((........))))..))). ( -34.30, z-score = -1.58, R) >droEre2.scaffold_4644 973005 85 + 2521924 -------AUGCCAAUCUUGU------------GGAUUGGUUUCCCUAUUUUCGGGGAUUGCA------UU-GCCAUUCUUUUCGCUAAUGACAAUCGCAAAUGUCUUGCGA -------..(((((((....------------.))))))).(((((......)))))(((((------((-((.(......).)).)))).)))((((((.....)))))) ( -23.90, z-score = -1.90, R) >droSec1.super_10 822380 110 + 3036183 AUACCAGAUGCCAAUCUUGUAUCUUGUGGGUUGGGUUGGCUUCCCUAUUUUCGGGGAUUGCAAAUGCAUU-GGCAUUCUUUCCGCUAAUGACAAUCGCAAAUGUCUUGCGA .........((((((((...(((.....))).)))))))).(((((......))))).......((((((-(((.........))))))).)).((((((.....)))))) ( -32.50, z-score = -1.24, R) >droSim1.chrX_random 857899 110 + 5698898 AUGCCAGAUGCCAAUCUGGUAUCUUGUGGGUUGGGUUGGCUUCCCUAUUUUCGAAGAUUGCGAAUGCAUU-GGCAUUCUUUCCGCUAAUGACAAUCGCAAAUGUCUUGCGA (((((((((....)))))))))...(((((..((.....))..)))))..(((((((((((((.((((((-(((.........))))))).)).)))))...))))).))) ( -38.20, z-score = -2.76, R) >consensus AUGCCAGAUGCCAAUCUUGUAUCUUGUGGGUUGGGUUGGCUUCCCUAUUUUCGAGGAUUGCAAAUGCAUU_GGCAUUCUUUCCGCUAAUGACAAUCGCAAAUGUCUUGCGA ......((.((((((((...(((.....))).)))))))).)).........(((((.(((...........))).))))).(((....((((........))))..))). (-21.62 = -21.50 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:51 2011