| Sequence ID | dm3.chrX |
|---|---|
| Location | 989,866 – 989,940 |
| Length | 74 |
| Max. P | 0.966158 |

| Location | 989,866 – 989,940 |
|---|---|
| Length | 74 |
| Sequences | 4 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 85.81 |
| Shannon entropy | 0.23023 |
| G+C content | 0.34566 |
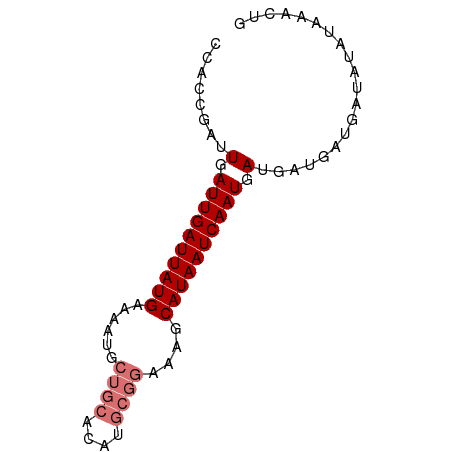
| Mean single sequence MFE | -14.82 |
| Consensus MFE | -11.82 |
| Energy contribution | -12.82 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
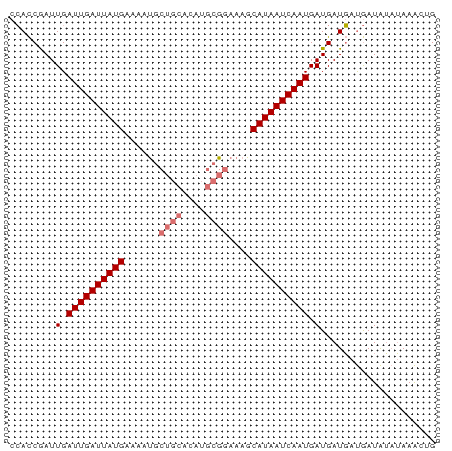
| Mean z-score | -2.40 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
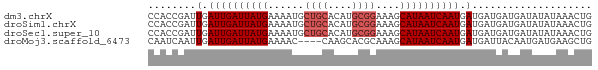
>dm3.chrX 989866 74 - 22422827 CCACCGAUUGAUUGAUUAUGAAAAUGCUGCACAUGCGGAAAGCAUAAUCAAUGAUGAUGAUGAUAUAUAAACUG .((.((..(.((((((((((......((((....))))....)))))))))).)...)).))............ ( -16.00, z-score = -2.89, R) >droSim1.chrX 797541 74 - 17042790 CCACCGAUUGAUUGAUUAUGAAAAUGCUGCACAUGCGGAAAGCAUAAUCAAUGAUGAUGAUGAUAUAUAAACUG .((.((..(.((((((((((......((((....))))....)))))))))).)...)).))............ ( -16.00, z-score = -2.89, R) >droSec1.super_10 817035 74 - 3036183 CCACCGAUUGAUUGAUUAUGAAAAUGCUGCACAUGCGGAAAGCAUAAUCAAUGAUGAUGAUGAUAUAUAAACUG .((.((..(.((((((((((......((((....))))....)))))))))).)...)).))............ ( -16.00, z-score = -2.89, R) >droMoj3.scaffold_6473 6938241 70 - 16943266 CAAUCAAUUGAUUGAUUAUGAAAAC----CAAGCACGCAAAGCAUAAUCAAUGAUGAUUACAAUGAUGAAGCUG .(((((..(.((((((((((.....----(......).....)))))))))).))))))............... ( -11.30, z-score = -0.95, R) >consensus CCACCGAUUGAUUGAUUAUGAAAAUGCUGCACAUGCGGAAAGCAUAAUCAAUGAUGAUGAUGAUAUAUAAACUG ........(.((((((((((......((((....))))....)))))))))).).................... (-11.82 = -12.82 + 1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:50 2011