| Sequence ID | dm3.chrX |
|---|---|
| Location | 950,834 – 951,008 |
| Length | 174 |
| Max. P | 0.968905 |

| Location | 950,834 – 950,945 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.59 |
| Shannon entropy | 0.07446 |
| G+C content | 0.36857 |
| Mean single sequence MFE | -17.73 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.69 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 950834 111 - 22422827 UGCAACUAUGUUGAUAAUUCUUGCUGUAACGCAUAAAUAAAGCCCAAAUUACAAAUUGGAUGCGAACGUAUGGAUUACUCGUACCUAAAACACAACCUCUCCCAAAAAGUG .(((.(((..(((.(((((...(((...............)))...))))))))..))).)))....((((((.....))))))........................... ( -13.66, z-score = 0.30, R) >droSim1.chrX 769715 109 - 17042790 UGCAACUAUGUUGGUAAUUCUUGCUGUAACGCAUAAAUAAAGCCCAAAUUACUAAUUGGAUGCGAACGUAUGAAUUACUCGUACCUGAAACACAACCAAUCCCAAAAUG-- .(((.(((.((((((((((...(((...............)))...))))))))))))).)))....((((((.....)))))).........................-- ( -19.76, z-score = -1.81, R) >droSec1.super_10 786650 111 - 3036183 UGCAACUAUGUUGGUAAUUCUUGCUGUAACGCAUAAAUAAAGCCCAAAUUACUAAUUGGAUGCGAACGUAUGAAUUACUCGUACCUGAAACACAACCAAUCCCAAAAAGUG .(((.(((.((((((((((...(((...............)))...))))))))))))).)))....((((((.....))))))........................... ( -19.76, z-score = -1.39, R) >consensus UGCAACUAUGUUGGUAAUUCUUGCUGUAACGCAUAAAUAAAGCCCAAAUUACUAAUUGGAUGCGAACGUAUGAAUUACUCGUACCUGAAACACAACCAAUCCCAAAAAGUG .(((.(((.((((((((((...(((...............)))...))))))))))))).)))....((((((.....))))))........................... (-17.25 = -17.69 + 0.45)


| Location | 950,905 – 951,008 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 68.45 |
| Shannon entropy | 0.51628 |
| G+C content | 0.35428 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -11.26 |
| Energy contribution | -10.45 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.968905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 950905 103 + 22422827 UUAUUUAUGCGUUACAGCAAGAAUUAUCAACAUAGUUGCACUCAUUAUGGUCGAUAAUGCUCGUAACUUAUCGUAAGAUAAGUUCAACAGCACAUGCAGUAAU ..........(((((.(((...((((((..((((((.......))))))...))))))(((.((((((((((....))))))))..)))))...))).))))) ( -26.20, z-score = -3.15, R) >droSim1.chrX 769784 103 + 17042790 UUAUUUAUGCGUUACAGCAAGAAUUACCAACAUAGUUGCACUCAUUAUGGUCGAUAAUGCUCGUAACUUAUCGUAAGAUAAGUUAAACAGCACAUACAGUAAU .......(((......)))...(((((...((((((.......))))))........((((..(((((((((....)))))))))...))))......))))) ( -23.20, z-score = -2.61, R) >droSec1.super_10 786721 103 + 3036183 UUAUUUAUGCGUUACAGCAAGAAUUACCAACAUAGUUGCACUCAUUAUGGUCGAUAAUGCUCGUAACUUAUCGCAAGAUAAGUUAAACAGCACAUACAGUAAU .......(((......)))...(((((...((((((.......))))))........((((..(((((((((....)))))))))...))))......))))) ( -24.20, z-score = -3.00, R) >droVir3.scaffold_12726 1198845 81 - 2840439 ------GUAUAAUCAAGUUGGAAGAGGCUACA--GUUGCCCCAAGCGAAAUUUAGAUUUUUCGAGCUUUGUC-UGAAACAAGCUGAACAG------------- ------......(((((((.((((((.(((..--.((((.....))))....))).)))))).))))((((.-....))))..)))....------------- ( -13.90, z-score = 0.56, R) >consensus UUAUUUAUGCGUUACAGCAAGAAUUACCAACAUAGUUGCACUCAUUAUGGUCGAUAAUGCUCGUAACUUAUCGUAAGAUAAGUUAAACAGCACAUACAGUAAU ........(((((((.((.........((((...))))...........)).).))))))....((((((((....))))))))................... (-11.26 = -10.45 + -0.81)

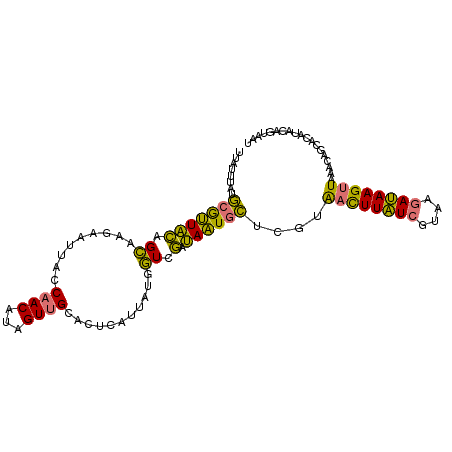
| Location | 950,905 – 951,008 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 68.45 |
| Shannon entropy | 0.51628 |
| G+C content | 0.35428 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -10.66 |
| Energy contribution | -13.78 |
| Covariance contribution | 3.13 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 950905 103 - 22422827 AUUACUGCAUGUGCUGUUGAACUUAUCUUACGAUAAGUUACGAGCAUUAUCGACCAUAAUGAGUGCAACUAUGUUGAUAAUUCUUGCUGUAACGCAUAAAUAA .........(((((.((((((((((((....)))))))).((((.(((((((((.(((.((....))..)))))))))))).))))...)))))))))..... ( -25.50, z-score = -2.43, R) >droSim1.chrX 769784 103 - 17042790 AUUACUGUAUGUGCUGUUUAACUUAUCUUACGAUAAGUUACGAGCAUUAUCGACCAUAAUGAGUGCAACUAUGUUGGUAAUUCUUGCUGUAACGCAUAAAUAA .....(((.(((((.((((((((((((....)))))))))((((.(((((((((.(((.((....))..)))))))))))).))))....)))))))).))). ( -24.60, z-score = -1.98, R) >droSec1.super_10 786721 103 - 3036183 AUUACUGUAUGUGCUGUUUAACUUAUCUUGCGAUAAGUUACGAGCAUUAUCGACCAUAAUGAGUGCAACUAUGUUGGUAAUUCUUGCUGUAACGCAUAAAUAA .....(((.(((((.((((((((((((....)))))))))((((.(((((((((.(((.((....))..)))))))))))).))))....)))))))).))). ( -24.60, z-score = -1.63, R) >droVir3.scaffold_12726 1198845 81 + 2840439 -------------CUGUUCAGCUUGUUUCA-GACAAAGCUCGAAAAAUCUAAAUUUCGCUUGGGGCAAC--UGUAGCCUCUUCCAACUUGAUUAUAC------ -------------...(((((((((((...-))).))))).))).((((............(((((...--....))))).........))))....------ ( -13.60, z-score = 0.02, R) >consensus AUUACUGUAUGUGCUGUUUAACUUAUCUUACGAUAAGUUACGAGCAUUAUCGACCAUAAUGAGUGCAACUAUGUUGGUAAUUCUUGCUGUAACGCAUAAAUAA .........(((((.((((((((((((....)))))))).((((.((((((((((((.....))).......))))))))).))))...)))))))))..... (-10.66 = -13.78 + 3.13)

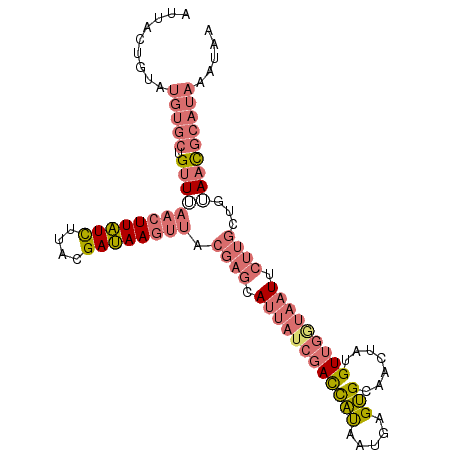
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:46 2011