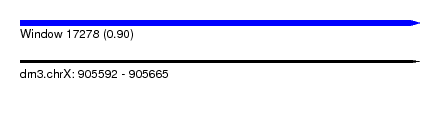
| Sequence ID | dm3.chrX |
|---|---|
| Location | 905,592 – 905,665 |
| Length | 73 |
| Max. P | 0.904304 |

| Location | 905,592 – 905,665 |
|---|---|
| Length | 73 |
| Sequences | 7 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 55.66 |
| Shannon entropy | 0.83080 |
| G+C content | 0.41349 |
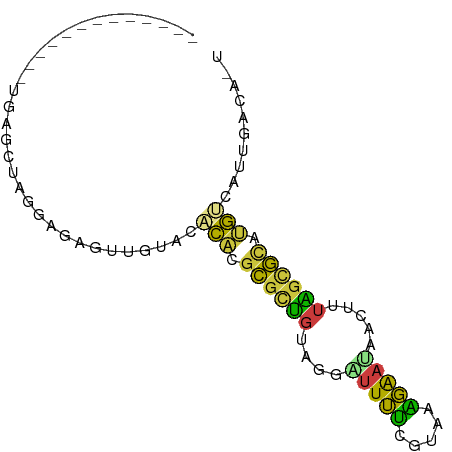
| Mean single sequence MFE | -17.47 |
| Consensus MFE | -8.02 |
| Energy contribution | -6.64 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 2.07 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
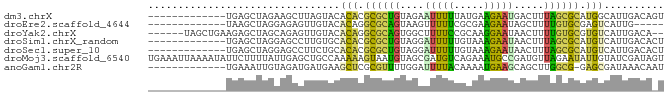
>dm3.chrX 905592 73 + 22422827 -------------UGAGCUAGAAGCUUAGUACACACGCGCUGUAGAAUUUUUAUGAAGAAUGACUUUAGCGCAUGGCAUUGACAGU -------------(((((.....))))).....((.((((((.((.(((((.....)))))..)).)))))).))........... ( -16.50, z-score = -0.58, R) >droEre2.scaffold_4644 885826 68 + 2521924 -------------UAAGCUAGGAGAGUUGUACACAGGCGCAGUAAGUUUUUCGCGAAGAAUAGCUUUUGUGCGAGUCAUUG----- -------------..........((.(((((((.((((.......((((((....)))))).)))).))))))).))....----- ( -15.40, z-score = -0.17, R) >droYak2.chrX 828048 78 + 21770863 ------UAGCUGAAGAGCUAGCAGAGUUGUACACAGGCGCAGUGGCUUUUCCGCAAGGAAUAACUUUUGUGCGUGUCAUUGACA-- ------(((((....))))).....((((...(((.((((((.((...((((....))))...)).)))))).)))...)))).-- ( -26.60, z-score = -1.80, R) >droSim1.chrX_random 849126 73 + 5698898 -------------UGAGCUAGGAGCCUUGUGCACACGCGCUGUAGGAUUUUUGUAAAGAAUAACUUUAGCGCAUGUCAUUGACACU -------------...((.((....))...))....((((((.((...((((....))))...)).)))))).((((...)))).. ( -16.40, z-score = 0.18, R) >droSec1.super_10 748504 73 + 3036183 -------------UGAGCUAGGAGCCUUCUGCACACGCGCUGUAGGAUUUUUGUAAAGAAUAACUUUAGCGCAUGUCAUUGACACU -------------...((.((....))...))....((((((.((...((((....))))...)).)))))).((((...)))).. ( -16.40, z-score = -0.02, R) >droMoj3.scaffold_6540 13412568 86 + 34148556 UGAAAUUAAAAUAUUCUUUUAUUGAGCUGCCAAAAAGUAAUGUAGCGAUGUCAGAAAUGCCGAUGUUAGAAUAUUGUAUCGAUAGU .........((((((((..(((((.(((((...........))))).....((....)).)))))..))))))))........... ( -16.30, z-score = -1.87, R) >anoGam1.chr2R 5564706 72 - 62725911 -------------UGAAAUUGUAGAUGAUGAAGCUCGCGUUUUGGAUUUUACAAAAUGAAGCAGCUUGGCG-GAGCGAUAAACAAU -------------....(((((...((.(.(((((..(((((((.......)))))))....))))).)))-..)))))....... ( -14.70, z-score = -0.71, R) >consensus _____________UGAGCUAGGAGAGUUGUACACACGCGCUGUAGGAUUUUCGUAAAGAAUAACUUUAGCGCAUGUCAUUGACA_U ................................(((.((((((....(((((.....))))).....)))))).))).......... ( -8.02 = -6.64 + -1.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:41 2011