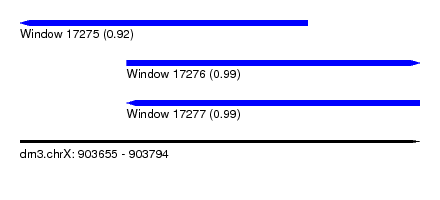
| Sequence ID | dm3.chrX |
|---|---|
| Location | 903,655 – 903,794 |
| Length | 139 |
| Max. P | 0.992634 |

| Location | 903,655 – 903,755 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Shannon entropy | 0.31628 |
| G+C content | 0.33011 |
| Mean single sequence MFE | -22.56 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
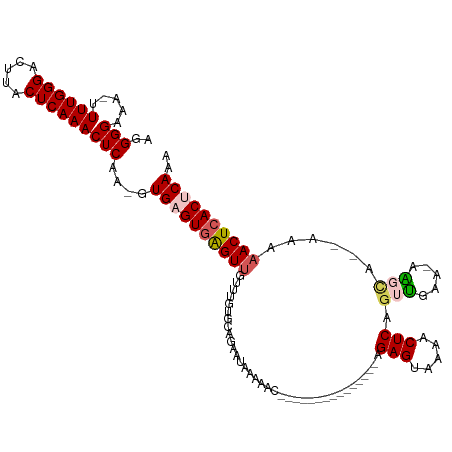
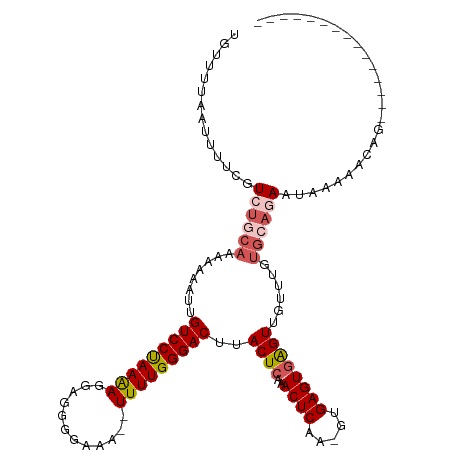
>dm3.chrX 903655 100 - 22422827 AGGGGAAA-UUUUGGGACUUACUCAAACUCAA-GUGAGUGAGUUUAUUGUGCAGAAUAAAAUC---------------AGAGUAAAACUCAGUAGGA-AUGCA--AAAAACUCACUCAAA ..(((...-.((((((.....)))))))))..-.(((((((((((....((((((......))---------------.(((.....))).......-.))))--..))))))))))).. ( -26.30, z-score = -2.96, R) >droSim1.chrX_random 847169 100 - 5698898 AGGGGAAA-UUUUGGGACUUACUCAAACUCAA-GUGAGUGAGUUGUUUGUGCAGAAUAAAAAC---------------AGAGUAAAACUCAGUUGAA-AAGCA--AAAAACUCACACAAA ........-.((((.((((((((((.......-.))))))))))...((((.((.........---------------.(((.....))).(((...-.))).--.....)))))))))) ( -22.10, z-score = -1.22, R) >droSec1.super_10 746718 99 - 3036183 AGGGGAAA-UUUUGGGAC-UACUCAAACUCAA-GUGAGUGAGUUGUUUGUGCAGAAUAAAAAC---------------AGAGUAAAACUCAGUUGAA-AAGCA--AAAAACUCACACAAA ..(((...-.((((((..-..)))))))))..-(((((((((((.(((((...........))---------------)))....))))).(((...-.))).--....))))))..... ( -22.00, z-score = -1.40, R) >droYak2.chrX 826249 97 - 21770863 AGGGGAAAUUUUUGGGACUUACUCAAACUCAAAGUGAGUAAGUAAA------AAAUUAAAAAU---------------AGAGUAAAACUCAGUUGAAUAGUGA--AAAAACUCACUCAAA .(((..(((((((...(((((((((.........))))))))).))------)))))......---------------.((((.....(((.(.....).)))--....)))).)))... ( -20.50, z-score = -2.60, R) >droEre2.scaffold_4644 883960 120 - 2521924 AGGGGAAAUUUUUGGGACGUACUCAAACUCAAUGUGAGUGGGUUGUUGGUGCAAAAAAAAAACAAAGAAAUAAAAAAUAGAGUAAAACUCAGUAUAAUAUCUAACAAAAACUCACUCAAA ..(((.....((((((.....)))))))))....(((((((((((((((................(....)........(((.....)))..........))))))...))))))))).. ( -21.90, z-score = -2.01, R) >consensus AGGGGAAA_UUUUGGGACUUACUCAAACUCAA_GUGAGUGAGUUGUUUGUGCAGAAUAAAAAC_______________AGAGUAAAACUCAGUUGAA_AAGCA__AAAAACUCACUCAAA ..(((.....((((((.....)))))))))....((((((((((...................................(((.....))).(((.....)))......)))))))))).. (-15.94 = -16.50 + 0.56)


| Location | 903,692 – 903,794 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.48 |
| Shannon entropy | 0.38555 |
| G+C content | 0.34162 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -14.41 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 903692 102 + 22422827 ---------------CUGAUUUUAUUCUGCACAAUAAACUCACUCAC-UUGAGUUUGAGUAAGUCCCAAAA--UUUCCCCUUCUUUUAGGACAAUUUUUUUGCAGACGAAACUUAAAACG ---------------.(((.(((..((((((...((((((((.....-.)))))))).....((((.((((--..........)))).))))........))))))..))).)))..... ( -20.30, z-score = -2.85, R) >droSim1.chrX_random 847206 102 + 5698898 ---------------CUGUUUUUAUUCUGCACAAACAACUCACUCAC-UUGAGUUUGAGUAAGUCCCAAAA--UUUCCCCUCCUUUUAGGACAAUAUUUUUGUAGACGAAAAUUAAAACA ---------------..((((((..((((((.(((..((((((((..-..))))..))))..((((.((((--..........)))).))))....))).)))))).))))))....... ( -19.40, z-score = -2.22, R) >droSec1.super_10 746755 101 + 3036183 ---------------CUGUUUUUAUUCUGCACAAACAACUCACUCAC-UUGAGUUUGAGUA-GUCCCAAAA--UUUCCCCUCCUUUUAGGACAAUAUUUUUGCAGACCAAAAUUAAAACA ---------------..(((((.((((((((.(((..((((((((..-..))))..)))).-((((.((((--..........)))).))))....))).)))))).....)).))))). ( -20.80, z-score = -3.30, R) >droYak2.chrX 826287 99 + 21770863 ---------------------CUAUUUUUAAUUUUUUACUUACUCACUUUGAGUUUGAGUAAGUCCCAAAAAUUUCCCCUCCUUCUUGGGACAAUUUUUUGGCGGACGCAAAUGAAAACA ---------------------...((((((.....((((((((((.....))))..))))))(((((.((((((..(((........)))..))))))...).)))).....)))))).. ( -21.60, z-score = -2.17, R) >droEre2.scaffold_4644 884000 120 + 2521924 CUAUUUUUUAUUUCUUUGUUUUUUUUUUGCACCAACAACCCACUCACAUUGAGUUUGAGUACGUCCCAAAAAUUUCCCCUCCCUCUUGGGACAAUUUUUUUGCAGACACAAAUUAAAACA ................((((((...((((......(((...((((.....))))))).....(((.((((((.......((((....)))).....))))))..))).))))..)))))) ( -19.00, z-score = -2.02, R) >consensus _______________CUGUUUUUAUUCUGCACAAACAACUCACUCAC_UUGAGUUUGAGUAAGUCCCAAAA__UUUCCCCUCCUUUUAGGACAAUUUUUUUGCAGACGAAAAUUAAAACA .........................((((((......((((((((.....))))..))))..((((......................))))........)))))).............. (-14.41 = -14.53 + 0.12)

| Location | 903,692 – 903,794 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.48 |
| Shannon entropy | 0.38555 |
| G+C content | 0.34162 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
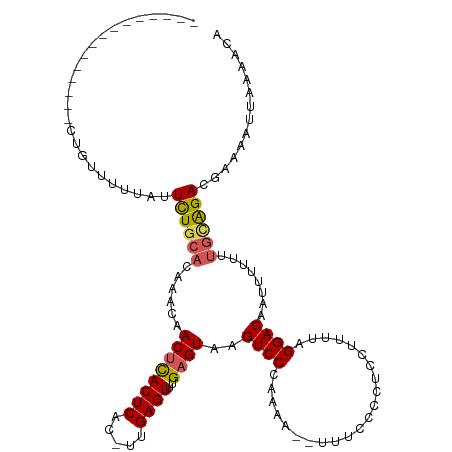
>dm3.chrX 903692 102 - 22422827 CGUUUUAAGUUUCGUCUGCAAAAAAAUUGUCCUAAAAGAAGGGGAAA--UUUUGGGACUUACUCAAACUCAA-GUGAGUGAGUUUAUUGUGCAGAAUAAAAUCAG--------------- ..............((((((..((((((.((((........))))))--)))).(((((((((((.......-.)))))))))))....))))))..........--------------- ( -26.10, z-score = -2.79, R) >droSim1.chrX_random 847206 102 - 5698898 UGUUUUAAUUUUCGUCUACAAAAAUAUUGUCCUAAAAGGAGGGGAAA--UUUUGGGACUUACUCAAACUCAA-GUGAGUGAGUUGUUUGUGCAGAAUAAAAACAG--------------- ((((((.(((((.((..(((((....((.((((......)))).)).--......((((((((((.......-.)))))))))).)))))))))))).)))))).--------------- ( -24.50, z-score = -2.05, R) >droSec1.super_10 746755 101 - 3036183 UGUUUUAAUUUUGGUCUGCAAAAAUAUUGUCCUAAAAGGAGGGGAAA--UUUUGGGAC-UACUCAAACUCAA-GUGAGUGAGUUGUUUGUGCAGAAUAAAAACAG--------------- ........(((((.((((((.(((((..(((((((((..........--)))))))))-.((((..((((..-..))))))))))))).)))))).)))))....--------------- ( -28.50, z-score = -3.32, R) >droYak2.chrX 826287 99 - 21770863 UGUUUUCAUUUGCGUCCGCCAAAAAAUUGUCCCAAGAAGGAGGGGAAAUUUUUGGGACUUACUCAAACUCAAAGUGAGUAAGUAAAAAAUUAAAAAUAG--------------------- ........(((((((((....(((((((..(((........)))..))))))).))))(((((((.........)))))))))))).............--------------------- ( -23.00, z-score = -2.04, R) >droEre2.scaffold_4644 884000 120 - 2521924 UGUUUUAAUUUGUGUCUGCAAAAAAAUUGUCCCAAGAGGGAGGGGAAAUUUUUGGGACGUACUCAAACUCAAUGUGAGUGGGUUGUUGGUGCAAAAAAAAAACAAAGAAAUAAAAAAUAG ((((((..((((..((.((((.......((((((((((..........))))))))))...(((..((((.....))))))))))).))..))))...))))))................ ( -29.90, z-score = -3.12, R) >consensus UGUUUUAAUUUUCGUCUGCAAAAAAAUUGUCCUAAAAGGAGGGGAAA__UUUUGGGACUUACUCAAACUCAA_GUGAGUGAGUUGUUUGUGCAGAAUAAAAACAG_______________ ..............((((((........(((((((((............)))))))))..((((..((((.....))))))))......))))))......................... (-17.42 = -18.38 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:40 2011