| Sequence ID | dm3.chrX |
|---|---|
| Location | 901,500 – 901,589 |
| Length | 89 |
| Max. P | 0.586871 |

| Location | 901,500 – 901,589 |
|---|---|
| Length | 89 |
| Sequences | 10 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 74.84 |
| Shannon entropy | 0.50119 |
| G+C content | 0.37916 |
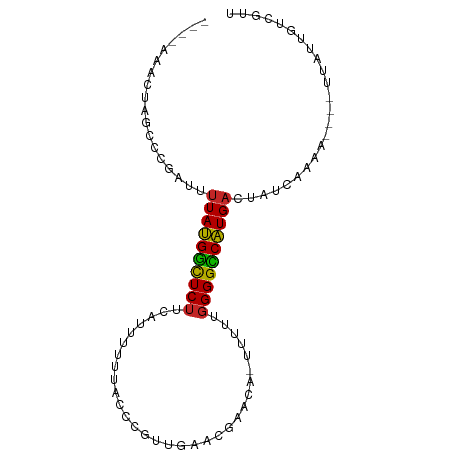
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -9.64 |
| Energy contribution | -9.09 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
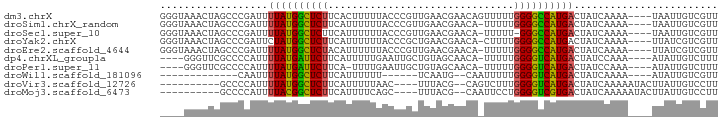
>dm3.chrX 901500 89 - 22422827 GGGUAAACUAGCCCGAUUUUAUGGCUCUUCACUUUUUACCCGUUGAACGAACAGUUUUUGGGGCCAUGACUAUCAAAA----UAAUUGUCGUU ((((......))))(((.(((((((.............((((..((((.....)))).)))))))))))..)))....----........... ( -21.91, z-score = -1.52, R) >droSim1.chrX_random 844504 88 - 5698898 GGGUAAACUAGCCCGAUUUUAUGGCUCUUCAUUUUUUACCCGUUGAACGAACA-UUUUUGGGGCCAUGACUAUCAAAA----UAAUUGUCGUU ((((......))))(((.(((((((((((((............))))((((..-..)))))))))))))..)))....----........... ( -20.90, z-score = -1.52, R) >droSec1.super_10 744588 87 - 3036183 GGGUAAACUAGCCCGAUUUUAUGGCUCUUCAUUUUUUACCCGUUGAACGAACA-UUUUU-GGGCCAUGACUAUCAAAA----UAAUUGUCGUU ((((......))))(((.(((((((((((((............))))......-.....-)))))))))..)))....----........... ( -18.70, z-score = -1.18, R) >droYak2.chrX 824085 88 - 21770863 GGGUAAACUAGCCCGAUUCUAUGGCUCUUCAUUUUUUACCCGCUGAACGAACA-CUUUUGGGGCCAUGACUAUCAAAA----UUAUCGUCGUU ((((......))))(((..((((((((((((............))))((((..-..))))))))))))...)))....----........... ( -18.60, z-score = -0.80, R) >droEre2.scaffold_4644 881865 88 - 2521924 GGGUAAACUAGCCCGAUUUUAUGGCUCUACAUUUUUUACCCGUUGAACGAACA-UUUUUGGGGCCAUGACUAUCAAAA----UUAUCGUCGUU ((((......))))(((.(((((((((((...........((.....))....-....)))))))))))..)))....----........... ( -20.81, z-score = -1.52, R) >dp4.chrXL_group1a 3893317 84 + 9151740 ----GGGUUCGCCCCAUUUUAUGAUUCUUCAUUUUUGAAUUGCUGUAGCAACA-UUUUUGGGGUCAUGACUAUCCAAA----AUAUUGUCUUU ----((((((((((((....(((....((((....))))((((....))))))-)...))))))...))..))))...----........... ( -18.70, z-score = -1.50, R) >droPer1.super_11 1001231 83 + 2846995 ----GGGUUCGCCCCAUUUUAUGAUUCUUCA-UUUUGAAUUGCUGUAGCAACA-UUUUUGGGGUCAUGACUAUCCAAA----AUAUUGUCUUU ----((((((((((((....(((.(((....-....)))((((....))))))-)...))))))...))..))))...----........... ( -17.70, z-score = -1.06, R) >droWil1.scaffold_181096 1743046 68 + 12416693 -------------CAAUUUUAUGGCUCUUCAUUUUUU------UCAAUG--CAAUUUUUGGGGUCAUGACUAUCAAAA----AUAUUGUCGUU -------------.....((((((((((.........------......--........)))))))))).........----........... ( -8.80, z-score = -0.79, R) >droVir3.scaffold_12726 1101943 77 + 2840439 ----------GCCCCAUUUUAUGGCUCUUCAUUUUUAAC----UUUACG--CAGUCUUUGGGGUCAUGACUAUCAAAAAUACUUAUUGUCCUU ----------((((((......(((((............----.....)--.))))..))))))...(((.................)))... ( -9.86, z-score = -0.05, R) >droMoj3.scaffold_6473 6778102 77 - 16943266 ----------GCCCCAUUUUACGGCUCUUCAUUUUCAGC----UUUACG--CAAUUCCUGGGGUCGUGACUAUCAAAAAUACUUAUUGUCCUU ----------........((((((((((.........((----.....)--).......))))))))))........................ ( -11.99, z-score = -0.67, R) >consensus ____AAACUAGCCCGAUUUUAUGGCUCUUCAUUUUUUACCCGUUGAACGAACA_UUUUUGGGGCCAUGACUAUCAAAA____UUAUUGUCGUU ..................((((((((((...............................))))))))))........................ ( -9.64 = -9.09 + -0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:36 2011