
| Sequence ID | dm3.chrX |
|---|---|
| Location | 894,605 – 894,701 |
| Length | 96 |
| Max. P | 0.895254 |

| Location | 894,605 – 894,701 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.32 |
| Shannon entropy | 0.51748 |
| G+C content | 0.48597 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -9.12 |
| Energy contribution | -9.77 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 894605 96 + 22422827 -------ACCCUGAACCAGACCAAGCCUCCC-CCGACCACCC-CCGCAAGAUAUAUGUUGACAGCUUGUGAAAAUAUGAGCAGCAUUUUGCUGCUGCCAGCCAAA -------...(((.......((((((.....-.((((.....-.(....)......))))...))))).)........((((((.....))))))..)))..... ( -19.10, z-score = -2.33, R) >droSim1.chrX 730759 96 + 17042790 -------ACCCUGAACCAGCCCAAACCUCCC-CCGACCACCC-CCGCAAGAUAUAUGUUGACAGCUUUUGAAAAUAUGAGCAGCAUUUUGCUGCUGCCAGCCAAA -------...(((..................-.((((.....-.(....)......))))..................((((((.....))))))..)))..... ( -14.80, z-score = -1.34, R) >droSec1.super_10 737580 96 + 3036183 -------ACCCUGAACCAGCCCAAACCUCCC-CCGACCACCC-CCGCAAGAUAUAUGUUGACAGCUUGUGAAAAUAUGAGCAGCAUUUUGCUGCUGCCAGCCAAA -------...(((..................-..........-.((((((.....((....)).))))))........((((((.....))))))..)))..... ( -17.90, z-score = -2.18, R) >droYak2.chrX 816362 75 + 21770863 ----------------------------CCC-CCCACCUCCC-CCGCAAGAUAUAUGUUGGCAGCUUGUGAAAAUAUGAGCAGCAUUUUGCUGCUGCCAGCCAAA ----------------------------...-..........-.(....)......(((((((((.(((....)))..(((((....)))))))))))))).... ( -21.30, z-score = -2.72, R) >droEre2.scaffold_4644 871250 79 + 2521924 -------------------------CCACCCCCCAAUCUCCC-CCGCAAGAUAUAUGUUGACAGCUUGUGAAAAUAUGAGCAGCAUUUUGCUGCUGCCAGCCAAA -------------------------.................-.((((((.....((....)).))))))......((.((((((......))))))))...... ( -16.50, z-score = -1.97, R) >droAna3.scaffold_12929 664901 105 + 3277472 UCCCUGGCGUAGUCCUGUUUUCUAAGGAUAUGCCAGCCACCCACCGUAAGAUAUAUGUUGACAGCUUUUGAAAAUAUGAGCAGCAUUUUGCUGCUGCCAGCCAAA ...(((((....((.(((((((.((((...((.((((.......(....)......)))).)).)))).))))))).))(((((.....))))).)))))..... ( -30.12, z-score = -2.40, R) >droWil1.scaffold_181096 1728247 88 - 12416693 ----------------UUGCUCACAUUUUCUGCCCUUCUCCC-UUGGAAGAUAUAUGUUGACAGCUUGGAAAAAUAUGAGCAGCAUUUUGCUAGUGCCAGCCAAA ----------------.((((((..((((((((.(((((...-..))))).....((....))))..))))))...))))))(((((.....)))))........ ( -18.60, z-score = 0.09, R) >droMoj3.scaffold_6473 6766948 72 + 16943266 ---------------------------------CCCCCACCCACAACAAGAUAUAUGUUGACAGCUUGUGAAAAUAUGAGCAACGUUUUGCUGCUACCAGCCAAA ---------------------------------..........(((((.......)))))...(((..((....))..)))........((((....)))).... ( -11.90, z-score = -1.29, R) >consensus __________________G__CAAACCUCCC_CCCACCACCC_CCGCAAGAUAUAUGUUGACAGCUUGUGAAAAUAUGAGCAGCAUUUUGCUGCUGCCAGCCAAA ...................................................(((((.((.((.....)).)).)))))((((((.....)))))).......... ( -9.12 = -9.77 + 0.66)

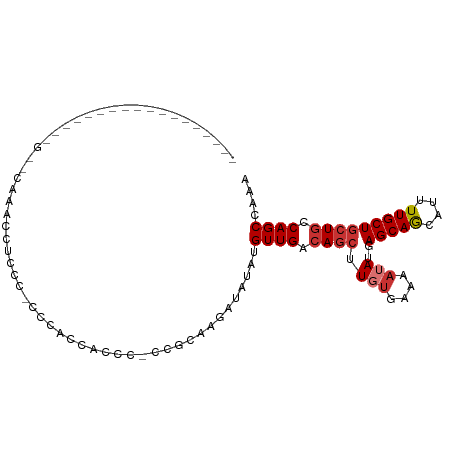
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:35 2011