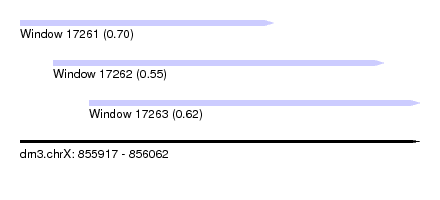
| Sequence ID | dm3.chrX |
|---|---|
| Location | 855,917 – 856,062 |
| Length | 145 |
| Max. P | 0.698017 |

| Location | 855,917 – 856,009 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.35 |
| Shannon entropy | 0.23173 |
| G+C content | 0.55831 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -26.39 |
| Energy contribution | -26.95 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.698017 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 855917 92 + 22422827 ----------------CUGCCUUCCCUUUCCCUUCCUGCUUUGUCCUGGACUGUACUGGCUGUCUGUCUUGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUU ----------------..((...((((.....(((((..((((..((((((......(((.....)))..))))))))))..))))).....))))...))....... ( -24.70, z-score = -0.18, R) >droEre2.scaffold_4644 831255 103 + 2521924 CUGCCCUUUUUCCCAUUCCCAUUCCCAUUCCCGUCCUGCUUUGUCCUGGGCUGU-CCGGCUGU----CUUGUCCAGCAAAAAAGGGACACCCGGGGCUUACUGCAAUU ....................................(((...((((((((.(((-((.((((.----......)))).......))))))))))))).....)))... ( -28.30, z-score = -0.65, R) >droSec1.super_10 697413 88 + 3036183 ----------------CUGCCUUCCCUUUCCCUUCCUGCUUUGUCCUGGGCUGUCCUGGCUGU----CUUGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUU ----------------....................(((...((((((((.((((((.((((.----......))))......)))))))))))))).....)))... ( -31.40, z-score = -1.81, R) >droSim1.chrX 690754 88 + 17042790 ----------------CUGCCUUCCCUUUCCCUUCCUGCUUUGUCCUGGGCUGUCCUGGCUGU----CUUGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUU ----------------....................(((...((((((((.((((((.((((.----......))))......)))))))))))))).....)))... ( -31.40, z-score = -1.81, R) >consensus ________________CUGCCUUCCCUUUCCCUUCCUGCUUUGUCCUGGGCUGUCCUGGCUGU____CUUGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUU ....................................(((...((((((((.((((((.((((...........))))......)))))))))))))).....)))... (-26.39 = -26.95 + 0.56)



| Location | 855,929 – 856,049 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.34 |
| Shannon entropy | 0.07437 |
| G+C content | 0.46482 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -31.14 |
| Energy contribution | -31.95 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 855929 120 + 22422827 UCCCUUCCUGCUUUGUCCUGGACUGUACUGGCUGUCUGUCUUGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUUUGUUGAAUUAAUAACAACACUCAGUCCUUUGAAAAUUGAA .........((...(((((((........(((.....))).(((((..........))))).)))))))..))..((((((((((.........))))..((((....)))))))))).. ( -27.00, z-score = 0.30, R) >droEre2.scaffold_4644 831283 115 + 2521924 UCCCGUCCUGCUUUGUCCUGGGCUGUCC-GGCUGUCU----UGUCCAGCAAAAAAGGGACACCCGGGGCUUACUGCAAUUUGUUGAAUUAAUAACAACACUCAGUGCUUUGGAAAUUGAA .....(((.((...((((((((.(((((-.((((...----....)))).......)))))))))))))..((((....((((((......))))))....))))))...)))....... ( -35.70, z-score = -1.54, R) >droSec1.super_10 697425 116 + 3036183 UCCCUUCCUGCUUUGUCCUGGGCUGUCCUGGCUGUCU----UGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUUUGUUGAAUUAAUAACAACACUCAGUGCUUUGAAAAUUGAA .........((...((((((((.((((((.((((...----....))))......))))))))))))))..))..((((((((((.........))))..((((....)))))))))).. ( -36.80, z-score = -2.12, R) >droSim1.chrX 690766 116 + 17042790 UCCCUUCCUGCUUUGUCCUGGGCUGUCCUGGCUGUCU----UGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUUUGUUGAAUUAAUAACAACACUCAGUGCUUUGAAAAUUGAA .........((...((((((((.((((((.((((...----....))))......))))))))))))))..))..((((((((((.........))))..((((....)))))))))).. ( -36.80, z-score = -2.12, R) >consensus UCCCUUCCUGCUUUGUCCUGGGCUGUCCUGGCUGUCU____UGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUUUGUUGAAUUAAUAACAACACUCAGUGCUUUGAAAAUUGAA .........((...((((((((.((((((.((((.(......)..))))......))))))))))))))..))..((((((((((.........))))..((((....)))))))))).. (-31.14 = -31.95 + 0.81)



| Location | 855,942 – 856,062 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.34 |
| Shannon entropy | 0.07437 |
| G+C content | 0.43486 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -32.12 |
| Energy contribution | -32.75 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 855942 120 + 22422827 UGUCCUGGACUGUACUGGCUGUCUGUCUUGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUUUGUUGAAUUAAUAACAACACUCAGUCCUUUGAAAAUUGAAUUUAUAGAGUGAA .(((((((........(((.....))).(((((..........))))).)))))))..........((((((......))))))(((((.((..((..(...)..))...)).))))).. ( -25.10, z-score = 0.93, R) >droEre2.scaffold_4644 831296 115 + 2521924 UGUCCUGGGCUGUCC-GGCUGUCU----UGUCCAGCAAAAAAGGGACACCCGGGGCUUACUGCAAUUUGUUGAAUUAAUAACAACACUCAGUGCUUUGGAAAUUGAAUUUAGAGAGUGAA .((((((((.(((((-.((((...----....)))).......))))))))))))).(((((....((((((......))))))....)))))(((((((.......)))))))...... ( -34.10, z-score = -1.26, R) >droSec1.super_10 697438 116 + 3036183 UGUCCUGGGCUGUCCUGGCUGUCU----UGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUUUGUUGAAUUAAUAACAACACUCAGUGCUUUGAAAAUUGAAUUUAGAGAGUGAA .((((((((.((((((.((((...----....))))......))))))))))))))..((((....((((((......))))))....)))).(((((((.......)))))))...... ( -37.10, z-score = -2.05, R) >droSim1.chrX 690779 116 + 17042790 UGUCCUGGGCUGUCCUGGCUGUCU----UGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUUUGUUGAAUUAAUAACAACACUCAGUGCUUUGAAAAUUGAAUUUAGAGAGUGAA .((((((((.((((((.((((...----....))))......))))))))))))))..((((....((((((......))))))....)))).(((((((.......)))))))...... ( -37.10, z-score = -2.05, R) >consensus UGUCCUGGGCUGUCCUGGCUGUCU____UGUCCAGCAAAAAAGGGACACCCGGGGCUUGCUGCAAUUUGUUGAAUUAAUAACAACACUCAGUGCUUUGAAAAUUGAAUUUAGAGAGUGAA .((((((((.((((((.((((.(......)..))))......))))))))))))))..((((....((((((......))))))....)))).(((((((.......)))))))...... (-32.12 = -32.75 + 0.62)

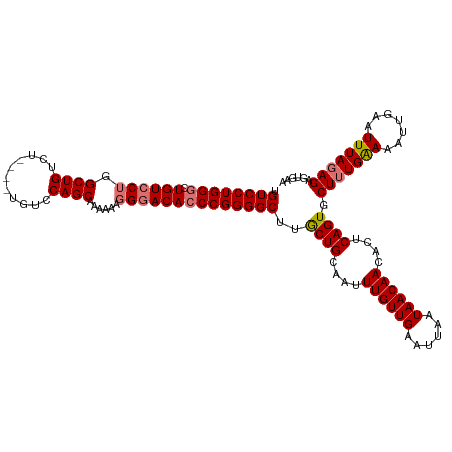
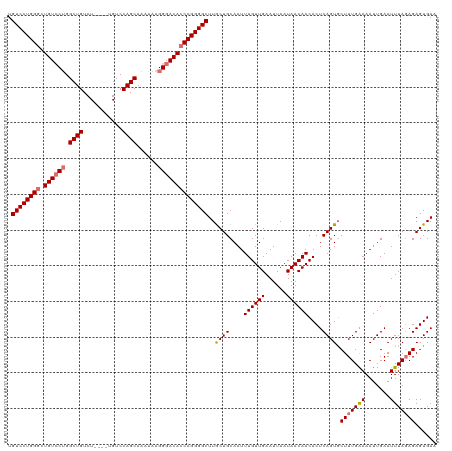
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:28 2011