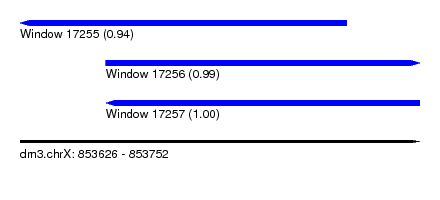
| Sequence ID | dm3.chrX |
|---|---|
| Location | 853,626 – 853,752 |
| Length | 126 |
| Max. P | 0.999412 |

| Location | 853,626 – 853,729 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 60.05 |
| Shannon entropy | 0.73203 |
| G+C content | 0.34880 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -7.04 |
| Energy contribution | -7.03 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.75 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
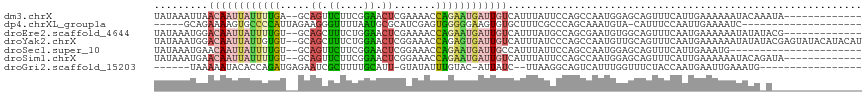
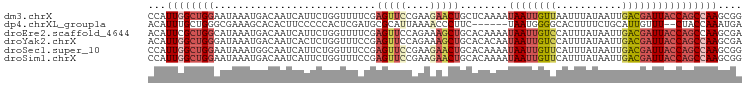
>dm3.chrX 853626 103 - 22422827 UAUAAAUUAACAAUUAUUUUGA--GCAGUUCUUCGGAACUCGAAAACCAGAAUGAUUGUCAUUUAUUCCAGCCAAUGGAGCAGUUUCAUUGAAAAAAUACAAAUA------------- .((((((..((((((((((((.--.((((((....))))).).....)))))))))))).))))))......((((((((...))))))))..............------------- ( -23.00, z-score = -3.13, R) >dp4.chrXL_group1a 3811832 92 + 9151740 -----GCAGAAAAGUGCCCCAUUAGAAGGGUUUUAAUGCGCAUCGAGUGGGGGAAGUGUGCUUUCGCCCAGCAAAUGUA-CAUUUCCAAUUGAAAAUC-------------------- -----(((......)))((((((.((.(.((......)).).)).))))))((((((((((.((.((...)).)).)))-)))))))...........-------------------- ( -27.70, z-score = -2.00, R) >droEre2.scaffold_4644 828404 103 - 2521924 UAUAAAUGGACAAUUAUUUUGU--GCAGCUUUCUGGAACUCGAAAACCAGAAUGAUUGUCAUUUAUGCCAGCGAAUGUGGCAGUUUCAAUGAAAAAAUAUAUACG------------- .(((((((.((((((((((((.--(....((((........)))).))))))))))))))))))))((((.......))))........................------------- ( -21.30, z-score = -1.12, R) >droYak2.chrX 771413 116 - 21770863 UAUAAAUGGACAAUUAUUGUGU--GCAGCUUUCUGGAACUCGGAAACCAGAGUGAUUGUCAUUUAUCCCAGCCAAUGUUGCAGUUUCAAUGAAAAAAUAUAUACGAGUAUACAUACAU .....(((.....((((((..(--(((((...((((.(((((....)..))))(((........))))))).....))))))....)))))).....(((((....)))))))).... ( -21.30, z-score = 0.06, R) >droSec1.super_10 695008 94 - 3036183 UAUAAAUGAACAAUUAUUUUGU--GCAGUUCUUCGGAACUCGGAAACCAGAAUGAUUGCCAUUUAUUCCAGCCAAUGGAGCAGUUUCAUUGAAAUG---------------------- .(((((((..(((((((((((.--(.(((((....))))))(....)))))))))))).)))))))......((((((((...)))))))).....---------------------- ( -26.40, z-score = -3.29, R) >droSim1.chrX 688316 103 - 17042790 UAUAAAUGAACAAUUAUUUUGU--GCAGUUCUUCGGAACUCGGAAACCAGAAUGAUUGUCAUUUAUUCCAGCCAAUGGAGCAGUUUCAUUGAAAAAAUACAGAUA------------- .((((((((.(((((((((((.--(.(((((....))))))(....))))))))))))))))))))......((((((((...))))))))..............------------- ( -29.50, z-score = -4.14, R) >droGri2.scaffold_15203 3902192 91 + 11997470 ------UAAAAAUACACCAGAUGAGAAUCGCUUUUGCAUU-GUAUAUUUGUAC-AUUAUC--UUAAGGCAGUCAUUUGGUUUCUACCAAUGAAUUGAAAUG----------------- ------.........(((((((((.....(((((.....(-((((....))))-).....--..)))))..))))))))).....................----------------- ( -15.80, z-score = -0.66, R) >consensus UAUAAAUGAACAAUUAUUUUGU__GCAGCUCUUUGGAACUCGGAAACCAGAAUGAUUGUCAUUUAUUCCAGCCAAUGGAGCAGUUUCAAUGAAAAAAUAUA_A_______________ .........((((((((((((.....((.((....)).)).......))))))))))))........................................................... ( -7.04 = -7.03 + -0.02)
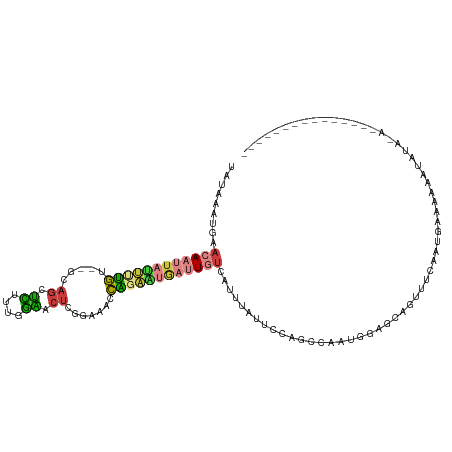
| Location | 853,653 – 853,752 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.57 |
| Shannon entropy | 0.45130 |
| G+C content | 0.40521 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -12.66 |
| Energy contribution | -14.08 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 853653 99 + 22422827 CCAUUGGCUGGAAUAAAUGACAAUCAUUCUGGUUUUCGAGUUCCGAAGAACUGCUCAAAAUAAUUGUUAAUUUAUAAUUGACGAUUACCAGCCAAGCGG ((.((((((((......(((.((((.....)))).)))(((((....)))))........(((((((((((.....)))))))))))))))))))..)) ( -29.50, z-score = -4.11, R) >dp4.chrXL_group1a 3811851 91 - 9151740 ACAUUUGCUGGGCGAAAGCACACUUCCCCCACUCGAUGCGCAUUAAAACCCUUC------UAAUGGGGCACUUUUCUGCAUUGUUU--CUACCAAAUGA .((((((.(((((....))..............(((((((........((((..------....))))........)))))))...--))).)))))). ( -23.99, z-score = -2.49, R) >droEre2.scaffold_4644 828431 99 + 2521924 ACAUUCGCUGGCAUAAAUGACAAUCAUUCUGGUUUUCGAGUUCCAGAAAGCUGCACAAAAUAAUUGUCCAUUUAUAAUUGACGAUUACCAGCCAAGCGA ....((((((((...((((.....)))).((((..(((.((..(((....))).))....(((((((......))))))).)))..))))))).))))) ( -24.60, z-score = -2.61, R) >droYak2.chrX 771453 99 + 21770863 ACAUUGGCUGGGAUAAAUGACAAUCACUCUGGUUUCCGAGUUCCAGAAAGCUGCACACAAUAAUUGUCCAUUUAUAAUUGACGAUUACCAGCCAAGCGA ...((((((((................(((((..........))))).............((((((((.((.....)).)))))))))))))))).... ( -25.90, z-score = -2.25, R) >droSec1.super_10 695026 99 + 3036183 CCAUUGGCUGGAAUAAAUGGCAAUCAUUCUGGUUUCCGAGUUCCGAAGAACUGCACAAAAUAAUUGUUCAUUUAUAAUUGACGAUUACCAGCCAAGCGG ((.((((((((......(((.((((.....)))).)))(((((....)))))........(((((((.((........)))))))))))))))))..)) ( -28.20, z-score = -2.91, R) >droSim1.chrX 688343 99 + 17042790 CCAUUGGCUGGAAUAAAUGACAAUCAUUCUGGUUUCCGAGUUCCGAAGAACUGCACAAAAUAAUUGUUCAUUUAUAAUUGACGAUUACCAGCCAAGCGG ((.((((((((..(((((((((((.(((.((((.....(((((....))))))).)).))).)))).)))))))(((((...)))))))))))))..)) ( -27.10, z-score = -3.10, R) >consensus ACAUUGGCUGGAAUAAAUGACAAUCAUUCUGGUUUCCGAGUUCCAAAAAACUGCACAAAAUAAUUGUUCAUUUAUAAUUGACGAUUACCAGCCAAGCGA ...((((((((...........................(((((....)))))........((((((((...........)))))))))))))))).... (-12.66 = -14.08 + 1.42)
| Location | 853,653 – 853,752 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
| Shannon entropy | 0.45130 |
| G+C content | 0.40521 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -15.72 |
| Energy contribution | -17.89 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
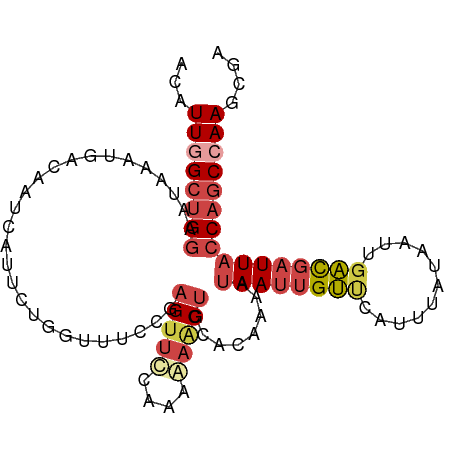
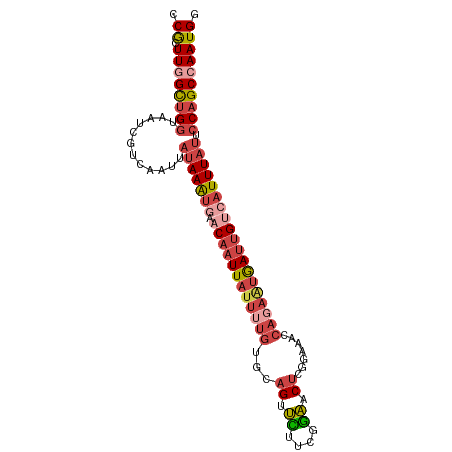
>dm3.chrX 853653 99 - 22422827 CCGCUUGGCUGGUAAUCGUCAAUUAUAAAUUAACAAUUAUUUUGAGCAGUUCUUCGGAACUCGAAAACCAGAAUGAUUGUCAUUUAUUCCAGCCAAUGG (((.((((((((............((((((..((((((((((((..((((((....))))).).....)))))))))))).)))))).))))))))))) ( -32.82, z-score = -5.37, R) >dp4.chrXL_group1a 3811851 91 + 9151740 UCAUUUGGUAG--AAACAAUGCAGAAAAGUGCCCCAUUA------GAAGGGUUUUAAUGCGCAUCGAGUGGGGGAAGUGUGCUUUCGCCCAGCAAAUGU ....((((..(--(((....(((.(....(.(((((((.------((.(.((......)).).)).))))))).)..).)))))))..))))....... ( -24.60, z-score = -0.72, R) >droEre2.scaffold_4644 828431 99 - 2521924 UCGCUUGGCUGGUAAUCGUCAAUUAUAAAUGGACAAUUAUUUUGUGCAGCUUUCUGGAACUCGAAAACCAGAAUGAUUGUCAUUUAUGCCAGCGAAUGU (((((.(((..((((..(.(((((((...((.((((.....)))).))....(((((..........)))))))))))).)..)))))))))))).... ( -28.10, z-score = -2.55, R) >droYak2.chrX 771453 99 - 21770863 UCGCUUGGCUGGUAAUCGUCAAUUAUAAAUGGACAAUUAUUGUGUGCAGCUUUCUGGAACUCGGAAACCAGAGUGAUUGUCAUUUAUCCCAGCCAAUGU ..((((((((((((((.....))))((((((.(((((((((.((.(....((((((.....))))))))).)))))))))))))))..)))))))).)) ( -29.40, z-score = -2.43, R) >droSec1.super_10 695026 99 - 3036183 CCGCUUGGCUGGUAAUCGUCAAUUAUAAAUGAACAAUUAUUUUGUGCAGUUCUUCGGAACUCGGAAACCAGAAUGAUUGCCAUUUAUUCCAGCCAAUGG (((.((((((((............(((((((..(((((((((((.(.(((((....))))))(....)))))))))))).))))))).))))))))))) ( -36.22, z-score = -5.38, R) >droSim1.chrX 688343 99 - 17042790 CCGCUUGGCUGGUAAUCGUCAAUUAUAAAUGAACAAUUAUUUUGUGCAGUUCUUCGGAACUCGGAAACCAGAAUGAUUGUCAUUUAUUCCAGCCAAUGG (((.((((((((............((((((((.(((((((((((.(.(((((....))))))(....)))))))))))))))))))).))))))))))) ( -39.32, z-score = -6.62, R) >consensus CCGCUUGGCUGGUAAUCGUCAAUUAUAAAUGAACAAUUAUUUUGUGCAGUUCUUCGGAACUCGGAAACCAGAAUGAUUGUCAUUUAUUCCAGCCAAUGG .((.((((((((............(((((((.((((((((((((...((.((....)).)).......))))))))))))))))))).)))))))))). (-15.72 = -17.89 + 2.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:24 2011