
| Sequence ID | dm3.chrX |
|---|---|
| Location | 848,406 – 848,497 |
| Length | 91 |
| Max. P | 0.917601 |

| Location | 848,406 – 848,497 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.29 |
| Shannon entropy | 0.41839 |
| G+C content | 0.42757 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -12.28 |
| Energy contribution | -12.72 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
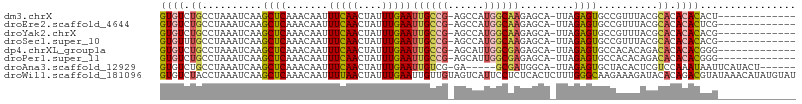
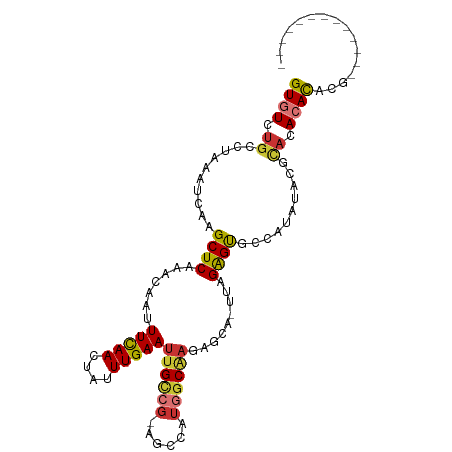
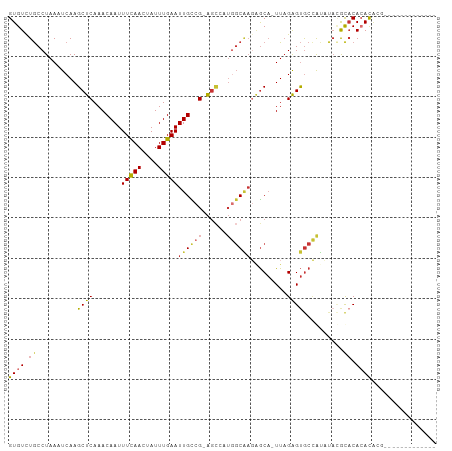
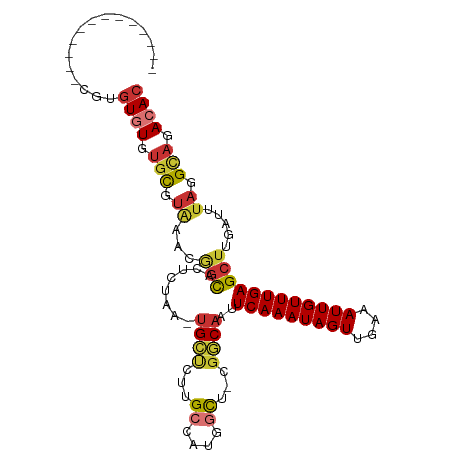
>dm3.chrX 848406 91 + 22422827 GUGUCUGCCUAAAUCAAGCUCAAACAAUUUCAACUAUUUGAAUUGCCG-AGCCAUGGCAAGAGCA-UUAGAGUGCCGUUUACGCACACACACU------------- ((((.(((.(((((...((((.......(((((....)))))((((((-.....)))))).....-...))))...))))).))).))))...------------- ( -24.50, z-score = -2.48, R) >droEre2.scaffold_4644 818765 91 + 2521924 GUGUCUGCCUAAAUCAAGCUCAAACAAUUUCAACUAUUUGAAUUGCCG-AGCCAUGGCAAGAGCA-UUAGAGUGCCGUUUACGCACACACUCG------------- ((((.(((.(((((...((((.......(((((....)))))((((((-.....)))))).....-...))))...))))).))).))))...------------- ( -24.00, z-score = -2.28, R) >droYak2.chrX 764823 91 + 21770863 GUGUCUGCCUAAAUCAAGCUCAAACAAUUUCAACUAUUUGAAUUGCCG-AGCCAUGGCAAGAGCA-UUAGAGUGCCGUUUACGCACACACACG------------- ((((.(((.(((((...((((.......(((((....)))))((((((-.....)))))).....-...))))...))))).))).))))...------------- ( -24.50, z-score = -2.50, R) >droSec1.super_10 685063 91 + 3036183 GUGUUUGCCUAAAUCAAGCUCAAACAAUUUCAACUAUUUGAAUUGCCG-AGCCAUGGCAAGAGCA-UUAGAGUGCCGUUUACGCACACACACG------------- ((((.(((.(((((...((((.......(((((....)))))((((((-.....)))))).....-...))))...))))).))).))))...------------- ( -24.50, z-score = -2.58, R) >dp4.chrXL_group1a 3806233 91 - 9151740 GUGUCUGCCUAAAUCAAGCUCAAACAAUUUCAACUAUUUGAAUUGCCG-AGCAUUGGCGAGAGCA-UUAGAGUGCCACACAGACACACACGGG------------- (((((((..........((((...(((((.(((....))))))))..)-)))..(((((....(.-...)..)))))..))))))).......------------- ( -24.60, z-score = -2.13, R) >droPer1.super_11 868691 91 - 2846995 GUGUCUGCCUAAAUCAAGCUCAAACAAUUUCAACUAUUUGAAUUGCCG-AGCAUUGGCGAGAGCA-UUAGAGUGCCACACAGACACACACGGG------------- (((((((..........((((...(((((.(((....))))))))..)-)))..(((((....(.-...)..)))))..))))))).......------------- ( -24.60, z-score = -2.13, R) >droAna3.scaffold_12929 608872 93 + 3277472 GUGUCUGCCUAAAUCAAGCUCAAACAAUUUCAACUAUUUGAAUUGUCG-GA-----GCGAUGGCA-UUAGAGUGCUACACUCGUCCAAAUAAUUCAUACU------ ((((.((((...(((..((((..((((((.(((....)))))))))..-))-----)))))))))-...(((((...))))).............)))).------ ( -22.80, z-score = -2.74, R) >droWil1.scaffold_181096 1663690 106 - 12416693 GUGUCUACCUAAAUCAAGCUCAAACAAUUUUAACUAUUUGAAUUGUUGUAGUCAUUCCUCUCACUCUUUGGGCAAGAAAGAUACACAGACGUAUAAACAUAUGUAU ((((((...........(((..((((((((.........))))))))..)))..(((..((((.....))))...)))))))))....((((((....)))))).. ( -17.20, z-score = -0.59, R) >consensus GUGUCUGCCUAAAUCAAGCUCAAACAAUUUCAACUAUUUGAAUUGCCG_AGCCAUGGCAAGAGCA_UUAGAGUGCCAUAUACGCACACACACG_____________ ((((.((..........((((.......(((((....)))))((((((......)))))).........))))..........)).))))................ (-12.28 = -12.72 + 0.44)
| Location | 848,406 – 848,497 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Shannon entropy | 0.41839 |
| G+C content | 0.42757 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -15.40 |
| Energy contribution | -16.04 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
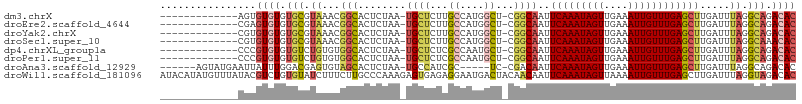
>dm3.chrX 848406 91 - 22422827 -------------AGUGUGUGUGCGUAAACGGCACUCUAA-UGCUCUUGCCAUGGCU-CGGCAAUUCAAAUAGUUGAAAUUGUUUGAGCUUGAUUUAGGCAGACAC -------------...((((.(((.((((.((((......-......))))..((((-(((((((((((....))).))))).)))))))...)))).))).)))) ( -28.20, z-score = -1.80, R) >droEre2.scaffold_4644 818765 91 - 2521924 -------------CGAGUGUGUGCGUAAACGGCACUCUAA-UGCUCUUGCCAUGGCU-CGGCAAUUCAAAUAGUUGAAAUUGUUUGAGCUUGAUUUAGGCAGACAC -------------...((((.(((.((((.((((......-......))))..((((-(((((((((((....))).))))).)))))))...)))).))).)))) ( -28.40, z-score = -1.96, R) >droYak2.chrX 764823 91 - 21770863 -------------CGUGUGUGUGCGUAAACGGCACUCUAA-UGCUCUUGCCAUGGCU-CGGCAAUUCAAAUAGUUGAAAUUGUUUGAGCUUGAUUUAGGCAGACAC -------------...((((.(((.((((.((((......-......))))..((((-(((((((((((....))).))))).)))))))...)))).))).)))) ( -28.20, z-score = -1.84, R) >droSec1.super_10 685063 91 - 3036183 -------------CGUGUGUGUGCGUAAACGGCACUCUAA-UGCUCUUGCCAUGGCU-CGGCAAUUCAAAUAGUUGAAAUUGUUUGAGCUUGAUUUAGGCAAACAC -------------.((((...(((.((((.((((......-......))))..((((-(((((((((((....))).))))).)))))))...)))).))).)))) ( -26.20, z-score = -1.51, R) >dp4.chrXL_group1a 3806233 91 + 9151740 -------------CCCGUGUGUGUCUGUGUGGCACUCUAA-UGCUCUCGCCAAUGCU-CGGCAAUUCAAAUAGUUGAAAUUGUUUGAGCUUGAUUUAGGCAGACAC -------------...((((.((((((((.((((......-))))..)))(((.(((-(((((((((((....))).))))).)))))))))....))))).)))) ( -28.60, z-score = -2.38, R) >droPer1.super_11 868691 91 + 2846995 -------------CCCGUGUGUGUCUGUGUGGCACUCUAA-UGCUCUCGCCAAUGCU-CGGCAAUUCAAAUAGUUGAAAUUGUUUGAGCUUGAUUUAGGCAGACAC -------------...((((.((((((((.((((......-))))..)))(((.(((-(((((((((((....))).))))).)))))))))....))))).)))) ( -28.60, z-score = -2.38, R) >droAna3.scaffold_12929 608872 93 - 3277472 ------AGUAUGAAUUAUUUGGACGAGUGUAGCACUCUAA-UGCCAUCGC-----UC-CGACAAUUCAAAUAGUUGAAAUUGUUUGAGCUUGAUUUAGGCAGACAC ------.((.(((((((..(((..(((((...)))))...-..)))..((-----((-.((((((((((....))).))))))).)))).))))))).))...... ( -24.30, z-score = -2.10, R) >droWil1.scaffold_181096 1663690 106 + 12416693 AUACAUAUGUUUAUACGUCUGUGUAUCUUUCUUGCCCAAAGAGUGAGAGGAAUGACUACAACAAUUCAAAUAGUUAAAAUUGUUUGAGCUUGAUUUAGGUAGACAC (((((.((((....)))).))))).((((.((..(.......)..))))))....((((..(((((((((((((....)))))))))).)))......)))).... ( -21.40, z-score = -0.70, R) >consensus _____________CGUGUGUGUGCGUAAACGGCACUCUAA_UGCUCUUGCCAUGGCU_CGGCAAUUCAAAUAGUUGAAAUUGUUUGAGCUUGAUUUAGGCAGACAC ................((((.(((.((...(((........((((...((....))...))))..(((((((((....)))))))))))).....)).))).)))) (-15.40 = -16.04 + 0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:21 2011